 |  | SWISS-MODEL Homology Modelling Report |
Model Building Report
This document lists the results for the homology modelling project "Q3XXF5-MurC-E-faecium" submitted to SWISS-MODEL workspace on Oct. 17, 2017, 5:59 p.m..The submitted primary amino acid sequence is given in Table T1.
If you use any results in your research, please cite the relevant publications:
Marco Biasini; Stefan Bienert; Andrew Waterhouse; Konstantin Arnold; Gabriel Studer; Tobias Schmidt; Florian Kiefer; Tiziano Gallo Cassarino; Martino Bertoni; Lorenza Bordoli; Torsten Schwede. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research (1 July 2014) 42 (W1): W252-W258; doi: 10.1093/nar/gku340.Arnold, K., Bordoli, L., Kopp, J. and Schwede, T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195-201.
Benkert, P., Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343-350
Results
The SWISS-MODEL template library (SMTL version 2017-10-11, PDB release 2017-10-06) was searched with Blast (Altschul et al., 1997) and HHBlits (Remmert, et al., 2011) for evolutionary related structures matching the target sequence in Table T1. For details on the template search, see Materials and Methods. Overall 5019 templates were found (Table T2).
Models
The following models were built (see Materials and Methods "Model Building"):
Model #01 | File | Built with | Oligo-State | Ligands | GMQE | QMEAN |
|---|---|---|---|---|---|---|
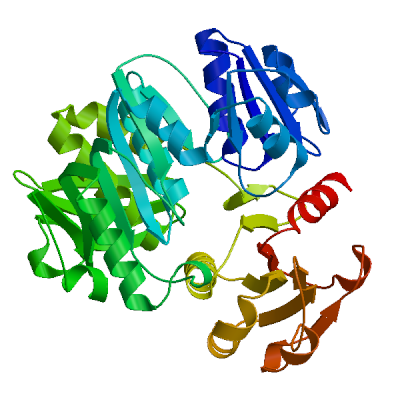 | PDB | ProMod3 Version 1.0.2. | MONOMER (matching prediction) | None | 0.66 | -4.80 |
|  |  |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 2f00.1.A | 28.70 | homo-dimer | HHblits | X-ray | 2.50Å | 0.34 | 5 - 435 | 0.97 | UDP-N-acetylmuramate--L-alanine ligase |
| Ligand | Added to Model | Description |
|---|---|---|
| MG | ✕ - Not biologically relevant. | MAGNESIUM ION |
| MG | ✕ - Not biologically relevant. | MAGNESIUM ION |
Target MENQNKLYHFVGIKGSGMSSLALVLHEQGLNVQGSDIEKYFFTQRDLEKANITI-LPFNADNVKPGMTIIAGNAFPDSHE
2f00.1.A ----VRHIHFVGIGGAGMGGIAEVLANEGYQISGSDLAPNPVTQ-QLMNLGATIYFNHRPENVRDASVVVVSSAISADNP
Target EIQRAKELGLEVIRYHDFIGHFIQNYTSIAVTGSHGKTSTTGLLSHVLSGVR-PTSYLIGD--GT---GHGDPQAEFFSF
2f00.1.A EIVAAHEARIPVIRRAEMLAELMRFRHGIAIAGTHGKTTTTAMVSSIYAEAGLDPTFVNGGLVKAAGVHARLGHGRYLIA
Target EACEYRRHFLAYSPDYAIMTNIDFDHPDYYT-SIDDVFTAFQTMAGQV--KKAIFAYGDDAYLRKLKA--NVPIYYYGVT
2f00.1.A EADESDASFLHLQPMVAIVTNIEADHMDTYQGDFENLKQTFINFLHNLPFYGRAVMCVDDPVIRELLPRVGRQTTTYGFS
Target ENDDIQARNIERTTSGSAFDVYHGDEFVGHFTVPAFGKHNILNALGVIAVAYFEKLDLKEVAEEMLTFPGVKRRFSEKIV
2f00.1.A EDADVRVEDYQQIGPQGHFTLLRQDKEPMRVTLNAPGRHNALNAAAAVAVATEEGIDDEAILRALESFQGTGRRFDFLGE
Target ---------ADM-TVVDDYAHHPAEIKATIDGARQKYPDKEIIAVFQPHTFTRTIALMDEFAEALDLADKVYLCDIFGSA
2f00.1.A FPLEPVNGKSGTAMLVDDYGHHPTEVDATIKAARAGWPDKNLVMLFQPHRFTRTRDLYDDFANVLTQVDTLLMLEVYPAG
Target REEQGNVKIEDLGAKIKKG--G---EVIKENN-VSPLL--DYHDAVVIFMGAGDVQKFEQAYEKL-LSSTTKNVL
2f00.1.A EAPIPGADSRSLCRTIRGRGKIDPILVPDPARVAEMLAPVLTGNDLILVQGAGNIGKIARSLAEIKLK-------
Model #02 | File | Built with | Oligo-State | Ligands | GMQE | QMEAN |
|---|---|---|---|---|---|---|
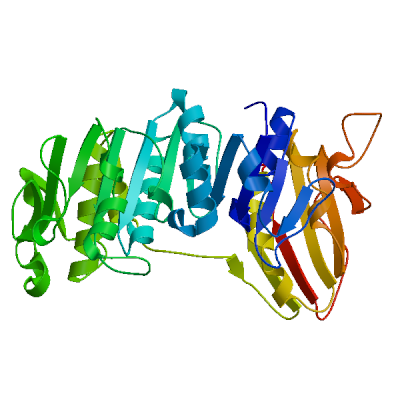 | PDB | ProMod3 Version 1.0.2. | MONOMER | None | 0.56 | -5.65 |
|  |  |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 1eeh.1.A | 17.50 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 3 - 429 | 0.90 | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE |
| Ligand | Added to Model | Description |
|---|---|---|
| UMA | ✕ - Binding site not conserved. | URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE |
Target MENQNKLYHFVGIKGSGMSSLALVLHEQGLNVQGSDIEKYFFTQRDLEKANITI-L-PFNADNVKPGMTIIAGNAFPDSH
1eeh.1.A --YQGKNVVIIGLGLTGLS-CVDFFLARGVTPRVMDTRMTPPGLD-KLPEAVERHTGSLNDEWLMAADLIVASPGIALAH
Target EEIQRAKELGLEVIRYHDFIGHFIQNYTSIAVTGSHGKTSTTGLLSHVLSGVRPTSYLIGD-GTGH---GDPQAEFFSFE
1eeh.1.A PSLSAAADAGIEIVGDIELFCRE-AQAPIVAITGSNGKSTVTTLVGEMAKAAGVNVGVGGNIGLPALMLLDDECELYVLE
Target ACEYRRHFL-AYSPDYAIMTNIDFDHPDYYT-SIDDVFTAFQTMAGQVKKAIFAYGDDAYLRKLKA-NVPIYYYGVTEND
1eeh.1.A LSSFQLETTSSLQAVAATILNVTEDHMDRYPFGLQQYRAAKLRIYEN-AKVCVVNADDALTMPIRGADERCVSFGVNM-G
Target DIQARNIERTTSGSAFDVYHGDE-FVGHFTVPAFGKHNILNALGVIAVAYFEKLDLKEVAEEMLTFPGVKRRFSEKIVAD
1eeh.1.A DYHLNHQQG--ETW-L--RVKGEKVLNVKEMKLSGQHNYTNALAALALADAAGLPRASSLKALTTFTGLPHRFEVVLEHN
Target M-TVVDDY-AHHPAEIKATIDGARQKYPDKEIIAVFQPHTFTRTIALMDEFAEALD-LADKVYLCDIFGSAREEQGNVKI
1eeh.1.A GVRWINDSKATNVGSTEAALNGLH--VD-GTLHLLLGGD--GKSADF-SPLARYLNGDNVRLYCFGRDG-----------
Target EDLGAKIKKGG-EVIKENN-VSPLL--DYHDAVVIFMGAG-DVQKFEQAYEKLLSSTTKNVL
1eeh.1.A AQLAALRPEVAEQTETMEQAMRLLAPRVQPGDMVLLSPACASLDQFK---------------
Model #04 | File | Built with | Oligo-State | Ligands | GMQE | QMEAN |
|---|---|---|---|---|---|---|
 | PDB | ProMod3 Version 1.0.2. | MONOMER | None | 0.51 | -5.06 |
|  |  |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 3zm5.1.A | 18.31 | monomer | HHblits | X-ray | 2.94Å | 0.29 | 55 - 433 | 0.82 | UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE |
| Ligand | Added to Model | Description |
|---|---|---|
| IGM | ✕ - Binding site not conserved. | 2,4-BIS(CHLORANYL)-N-[3-CYANO-6-[(4-HYDROXYPHENYL)METHYL]-5,7-DIHYDRO-4H-THIENO[2,3-C]PYRIDIN-2-YL]-5-MORPHOLIN-4-YLSULFONYL-BENZAMIDE |
Target MENQNKLYHFVGIKGSGMSSLALVLHEQGLNVQGSDIEKYFFTQRDLEKANITILPFNADNVKPGMTIIAGNAFPDSHEE
3zm5.1.A ------------------------------------------------------AEFDSRLIGTGDLFVPLKGARDGHDF
Target IQRAKELGLEV------------IRY---HDFIGHFIQ------NYTSIAVTGSHGKTSTTGLLSHVLSGVRPTSYLIGD
3zm5.1.A IETAFENGAAVTLSEKEVSNHPYILVDDVLTAFQSLASYYLEKTTVDVFAVTGSNGKTTTKDMLAHLLSTRYKTYKTQGN
Target -G----TGH----GDPQAEFFSFEACEYRR-H----FLAYSPDYAIMTNIDFDHPDYYTSIDDVFTAFQTMAGQV--KKA
3zm5.1.A YNNEIGLPYTVLHMPEGTEKLVLEMGQDHLGDIHLLSELARPKTAIVTLVGEAHLAFFKDRSEIAKGKMQIADGMASGSL
Target IFAYGDDAYLRKLKA-NVPIYYYGVTENDDIQARNIERTTSGSAFDVYHGDEFVGHFTVPAFGKHNILNALGVIAVAYFE
3zm5.1.A LLAPADPIVE-DYLPIDKKVVRFGQGAELEI--TDLVERKDSLTFKANFLEQ---ALDLPVTGKYNATNAMIASYVALQE
Target KLDLKEVAEEMLTFPGVKRRFSEKIVADM-TVVDD-YAHHPAEIKATIDGARQKY--PDKEIIAVFQPH--TFTRTIALM
3zm5.1.A GVSEEQIRLAFQHLELTRNRTEWKKAANGADILSDVYNANPTAMKLILETFSAIPANEGGKKIAVLADMKELGDQSVQLH
Target DEFAEALD-L-ADKVYLCDIFGSAREEQGNVKIEDLGAKIKKGG--EVIK------ENN-VSPLL--DYHDAVVIFMGAG
3zm5.1.A NQMILSLSPDVLDIVIFYGEDIA------QL-AQLASQMFPIGHVYYFKKTEDQDQFEDLVKQVKESLGAHDQILLKGSN
Target D--VQKFEQAYEKLLSSTTKNVL
3zm5.1.A SMNLAKLVESLE-----------
Model #03 | File | Built with | Oligo-State | Ligands | GMQE | QMEAN |
|---|---|---|---|---|---|---|
 | PDB | ProMod3 Version 1.0.2. | MONOMER | None | 0.50 | -5.86 |
|  |  |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 4buc.1.A | 14.32 | monomer | HHblits | X-ray | 2.17Å | 0.27 | 6 - 426 | 0.86 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| Ligand | Added to Model | Description |
|---|---|---|
| CL | ✕ - Not biologically relevant. | CHLORIDE ION |
| CL | ✕ - Not biologically relevant. | CHLORIDE ION |
| CL | ✕ - Not biologically relevant. | CHLORIDE ION |
| GOL | ✕ - Not biologically relevant. | GLYCEROL |
| GOL | ✕ - Not biologically relevant. | GLYCEROL |
| GOL | ✕ - Not biologically relevant. | GLYCEROL |
| NH4 | ✕ - Not biologically relevant. | AMMONIUM ION |
| PO3 | ✕ - Binding site not conserved. | PHOSPHITE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
| PO4 | ✕ - Not biologically relevant. | PHOSPHATE ION |
Target MENQNKLYHFVGIKGSGMSSLALVLHEQGLNVQGSDIEKYF-FTQRDLEKA-NITI-LP-FNADNVKPGMTIIAGNAFPD
4buc.1.A -----MKIGFLGFGKSNRSL-LKYLLNHQEAKFFVSEAKTLDGET-KKFLEEHSVEYEEGGHTEKLLDCDVVYVSPGIKP
Target SHEEIQRAKELGLEVIRYHDFI-GHFIQNYTSIAVTGSHGKTSTTGLLSHVLSGVRPTSYLIGD-GTGHG---DPQAEFF
4buc.1.A DTSMIELLSSRGVKLSTELQFFLDNVD-PKKVVGITGTDGKSTATALMYHVLSGRGFKTFLGGNFGTPAVEALEGEYDYY
Target SFEACEYRRHF-LAYSPDYAIMTNIDFDHPDYYTSIDDVFTAFQTM---AGQVKKAIFAYGDDAYLRKLKA-NVPIYYYG
4buc.1.A VLEMSSFQLFWSERPYLSNFLVLNISEDHLDWHSSFKEYVDSKLKPAFLQT-EGDLFVYNKHIERLRNLEGVRSRKIPFW
Target VTENDDIQARNIERTTSGSAFDVYHGDEFVGHFTVPAFGKHNIL---NALGVIAVAYFEKLDLKEVAEEMLTFPGVKRRF
4buc.1.A TDE--NF----A--TE-K-ELIV-------RGKKYTLPGNYPYQMRENILAVSVLYMEMFNELESFLELLRDFKPLPHRM
Target SEKIVADM-TVVDDY-AHHPAEIKATIDGARQKYPDKEIIAVFQPHTFTRTIALMDEFAEALD-LADKVYLCDIFGSARE
4buc.1.A EYLGQIDGRHFYNDSKATSTHAVLGALSNFD------KVVLIMCG--IGKKEN-YSLFVEKASPKLKHLIMFGEIS----
Target EQGNVKIEDLGAKIKK-GG-EVIKENN-VSPLL--DYHDAVVIFMGAGDVQKFEQAYEKLLSSTTKNVL
4buc.1.A -------KELAPFVGKIPHSIVENMEEAFEKAMEVSEKGDVILLSPGGASFDMYE--------------
Materials and Methods
Template Search
Template search with Blast and HHBlits has been performed against the SWISS-MODEL template library (SMTL, last update: 2017-10-11, last included PDB release: 2017-10-06).
The target sequence was searched with BLAST (Altschul et al., 1997) against the primary amino acid sequence contained in the SMTL. A total of 17 templates were found.
An initial HHblits profile has been built using the procedure outlined in (Remmert, et al., 2011), followed by 1 iteration of HHblits against NR20. The obtained profile has then be searched against all profiles of the SMTL. A total of 5012 templates were found.
Template Selection
For each identified template, the template's quality has been predicted from features of the target-template alignment. The templates with the highest quality have then been selected for model building.
Model Building
Models are built based on the target-template alignment using ProMod3. Coordinates which are conserved between the target and the template are copied from the template to the model. Insertions and deletions are remodelled using a fragment library. Side chains are then rebuilt. Finally, the geometry of the resulting model is regularized by using a force field. In case loop modelling with ProMod3 fails, an alternative model is built with PROMOD-II (Guex, et al., 1997).
Model Quality Estimation
The global and per-residue model quality has been assessed using the QMEAN scoring function (Benkert, et al., 2011) . For improved performance, weights of the individual QMEAN terms have been trained specifically for SWISS-MODEL.
Ligand Modelling
Ligands present in the template structure are transferred by homology to the model when the following criteria are met (Gallo -Casserino, to be published): (a) The ligands are annotated as biologically relevant in the template library, (b) the ligand is in contact with the model, (c) the ligand is not clashing with the protein, (d) the residues in contact with the ligand are conserved between the target and the template. If any of these four criteria is not satisfied, a certain ligand will not be included in the model. The model summary includes information on why and which ligand has not been included.
Oligomeric State Conservation
Homo-oligomeric structure of the target protein is predicted based on the analysis of pairwise interfaces of the identified template structures. For each relevant interface between polypeptide chains (interfaces with more than 10 residue-residue interactions), the QscoreOligomer (Mariani et al., 2011) is predicted from features such as similarity to target and frequency of observing this interface in the identified templates (Kiefer, Bertoni, Biasini, to be published). The prediction is performed with a random forest regressor using these features as input parameters to predict the probability of conservation for each interface. The QscoreOligomer of the whole complex is then calculated as the weight-averaged QscoreOligomer of the interfaces. The oligomeric state of the target is predicted to be the same as in the template when QscoreOligomer is predicted to be higher or equal to 0.5.
References
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res, 25, 3389-3402.
Remmert, M., Biegert, A., Hauser, A. and Soding, J. (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods, 9, 173-175.
Guex, N. and Peitsch, M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714-2723.
Sali, A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol, 234, 779-815.
Benkert, P., Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343-350.
Mariani, V., Kiefer, F., Schmidt, T., Haas, J. and Schwede, T. (2011) Assessment of template based protein structure predictions in CASP9. Proteins, 79 Suppl 10, 37-58.
Table T1:
Primary amino acid sequence for which templates were searched and models were built.
FIQNYTSIAVTGSHGKTSTTGLLSHVLSGVRPTSYLIGDGTGHGDPQAEFFSFEACEYRRHFLAYSPDYAIMTNIDFDHPDYYTSIDDVFTAFQTMAGQV
KKAIFAYGDDAYLRKLKANVPIYYYGVTENDDIQARNIERTTSGSAFDVYHGDEFVGHFTVPAFGKHNILNALGVIAVAYFEKLDLKEVAEEMLTFPGVK
RRFSEKIVADMTVVDDYAHHPAEIKATIDGARQKYPDKEIIAVFQPHTFTRTIALMDEFAEALDLADKVYLCDIFGSAREEQGNVKIEDLGAKIKKGGEV
IKENNVSPLLDYHDAVVIFMGAGDVQKFEQAYEKLLSSTTKNVL
Table T2:
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Coverage | Description |
|---|---|---|---|---|---|---|---|---|
| 3hn7.1.A | 33.65 | monomer | BLAST | X-ray | 1.65Å | 0.37 | 0.95 | UDP-N-acetylmuramate-L-alanine ligase |
| 2f00.1.A | 28.70 | homo-dimer | HHblits | X-ray | 2.50Å | 0.34 | 0.97 | UDP-N-acetylmuramate--L-alanine ligase |
| 4hv4.1.A | 29.47 | monomer | HHblits | X-ray | 2.25Å | 0.34 | 0.97 | UDP-N-acetylmuramate--L-alanine ligase |
| 4hv4.2.A | 29.47 | monomer | HHblits | X-ray | 2.25Å | 0.34 | 0.97 | UDP-N-acetylmuramate--L-alanine ligase |
| 2f00.1.A | 31.12 | homo-dimer | BLAST | X-ray | 2.50Å | 0.36 | 0.95 | UDP-N-acetylmuramate--L-alanine ligase |
| 1j6u.1.A | 34.87 | monomer | BLAST | X-ray | 2.30Å | 0.37 | 0.93 | UDP-N-acetylmuramate-alanine ligase MurC |
| 1p3d.1.A | 26.68 | monomer | HHblits | X-ray | 1.70Å | 0.34 | 0.97 | UDP-N-acetylmuramate--alanine ligase |
| 1gqq.1.A | 26.68 | homo-dimer | HHblits | X-ray | 3.10Å | 0.34 | 0.97 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1gqq.1.B | 26.68 | homo-dimer | HHblits | X-ray | 3.10Å | 0.34 | 0.97 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1p31.1.A | 26.68 | monomer | HHblits | X-ray | 1.85Å | 0.34 | 0.97 | UDP-N-acetylmuramate--alanine ligase |
| 1gqy.1.B | 26.68 | homo-dimer | HHblits | X-ray | 1.80Å | 0.34 | 0.97 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1gqy.1.A | 26.68 | homo-dimer | HHblits | X-ray | 1.80Å | 0.34 | 0.97 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 4hv4.1.A | 31.43 | monomer | BLAST | X-ray | 2.25Å | 0.36 | 0.95 | UDP-N-acetylmuramate--L-alanine ligase |
| 4hv4.2.A | 31.43 | monomer | BLAST | X-ray | 2.25Å | 0.36 | 0.95 | UDP-N-acetylmuramate--L-alanine ligase |
| 3hn7.1.A | 28.67 | monomer | HHblits | X-ray | 1.65Å | 0.33 | 0.97 | UDP-N-acetylmuramate-L-alanine ligase |
| 1p3d.1.A | 31.55 | monomer | BLAST | X-ray | 1.70Å | 0.36 | 0.93 | UDP-N-acetylmuramate--alanine ligase |
| 1gqq.1.A | 31.55 | homo-dimer | BLAST | X-ray | 3.10Å | 0.36 | 0.93 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1gqq.1.B | 31.55 | homo-dimer | BLAST | X-ray | 3.10Å | 0.36 | 0.93 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1p31.1.A | 31.55 | monomer | BLAST | X-ray | 1.85Å | 0.36 | 0.93 | UDP-N-acetylmuramate--alanine ligase |
| 1gqy.1.B | 31.55 | homo-dimer | BLAST | X-ray | 1.80Å | 0.36 | 0.93 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1gqy.1.A | 31.55 | homo-dimer | BLAST | X-ray | 1.80Å | 0.36 | 0.93 | UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE |
| 1j6u.1.A | 29.15 | monomer | HHblits | X-ray | 2.30Å | 0.34 | 0.95 | UDP-N-acetylmuramate-alanine ligase MurC |
| 5a5f.1.A | 17.46 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 2xpc.1.A | 17.21 | monomer | HHblits | X-ray | 1.49Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 1uag.1.A | 17.50 | monomer | HHblits | X-ray | 1.95Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYL-L-ALANINE/:D-GLUTAMATE LIGASE |
| 1eeh.1.A | 17.50 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE |
| 2uag.1.A | 17.50 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.90 | PROTEIN (UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE) |
| 1e0d.1.A | 17.50 | monomer | HHblits | X-ray | 2.40Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 5a5e.1.A | 17.50 | monomer | HHblits | X-ray | 1.84Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 3lk7.1.A | 19.44 | monomer | HHblits | X-ray | 1.50Å | 0.30 | 0.89 | UDP-N-acetylmuramoylalanine--D-glutamate ligase |
| 2y67.1.A | 17.54 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.90 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 4buc.1.A | 14.32 | monomer | HHblits | X-ray | 2.17Å | 0.27 | 0.86 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 4buc.2.A | 14.32 | monomer | HHblits | X-ray | 2.17Å | 0.27 | 0.86 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 4c12.1.A | 18.90 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.82 | UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--L-LYSINE LIGASE |
| 4bub.1.A | 23.69 | monomer | HHblits | X-ray | 2.90Å | 0.31 | 0.82 | UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--LD-LYSINE LIGASE |
| 4bub.2.A | 23.69 | monomer | HHblits | X-ray | 2.90Å | 0.31 | 0.82 | UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--LD-LYSINE LIGASE |
| 2xja.1.A | 19.40 | monomer | HHblits | X-ray | 3.00Å | 0.30 | 0.82 | UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE |
| 2wtz.1.A | 19.40 | monomer | HHblits | X-ray | 3.00Å | 0.30 | 0.82 | UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE |
| 1e8c.1.A | 21.15 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.82 | UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE |
| 1gg4.1.A | 18.68 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.82 | UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMYL-2,6-DIAMINOPIMELATE-D-ALANYL-D-ALANYL LIGASE |
| 3zm5.1.A | 18.31 | monomer | HHblits | X-ray | 2.94Å | 0.29 | 0.82 | UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE |
| 4qf5.1.A | 18.46 | monomer | HHblits | X-ray | 2.80Å | 0.29 | 0.82 | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
| 4qdi.1.A | 18.46 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.82 | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
| 3zl8.1.A | 18.96 | monomer | HHblits | X-ray | 1.65Å | 0.29 | 0.82 | UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE |
| 4ziy.1.A | 17.91 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.82 | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
| 4cvl.1.A | 19.10 | monomer | HHblits | X-ray | 2.98Å | 0.29 | 0.80 | UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE |
| 4cvm.1.A | 19.10 | monomer | HHblits | X-ray | 2.06Å | 0.29 | 0.80 | UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D- ALANINE LIGASE |
| 2am2.1.A | 18.16 | monomer | HHblits | X-ray | 2.80Å | 0.29 | 0.81 | UDP-N-acetylmuramoylalanine-D-glutamyl-lysine-D-alanyl-D-alanine ligase, MurF protein |
| 3eag.1.A | 31.72 | homo-dimer | BLAST | X-ray | 2.55Å | 0.36 | 0.65 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase |
| 3eag.1.B | 31.72 | homo-dimer | BLAST | X-ray | 2.55Å | 0.36 | 0.65 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase |
| 5vvw.1.A | 28.47 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.34 | 0.66 | UDP-N-acetylmuramate--L-alanine ligase |
| 5vvw.1.A | 31.49 | homo-tetramer | BLAST | X-ray | 2.30Å | 0.35 | 0.65 | UDP-N-acetylmuramate--L-alanine ligase |
| 3eag.1.A | 29.90 | homo-dimer | HHblits | X-ray | 2.55Å | 0.35 | 0.66 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase |
| 3eag.1.B | 29.90 | homo-dimer | HHblits | X-ray | 2.55Å | 0.35 | 0.66 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase |
| 2vor.1.A | 16.14 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.71 | FOLYLPOLYGLUTAMATE SYNTHASE PROTEIN FOLC |
| 1o5z.1.A | 13.52 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.72 | folylpolyglutamate synthase/dihydrofolate synthase |
| 2gc5.1.A | 15.34 | monomer | HHblits | X-ray | 1.85Å | 0.27 | 0.70 | Folylpolyglutamate synthase |
| 2gcb.1.A | 15.06 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.70 | Folylpolyglutamate synthase |
| 2gca.1.A | 15.76 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.70 | Folylpolyglutamate synthase |
| 1fgs.1.A | 15.38 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.70 | FOLYLPOLYGLUTAMATE SYNTHETASE |
| 1jbw.1.A | 15.38 | monomer | HHblits | X-ray | 1.85Å | 0.27 | 0.70 | FOLYLPOLYGLUTAMATE SYNTHASE |
| 1jbv.1.A | 15.38 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.70 | FOLYLPOLYGLUTAMATE SYNTHASE |
| 2gc6.1.A | 15.38 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.70 | Folylpolyglutamate synthase |
| 3nrs.1.A | 14.98 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.69 | Dihydrofolate:folylpolyglutamate synthetase |
| 3pyz.1.A | 14.98 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.69 | Bifunctional folylpolyglutamate synthase/dihydrofolate synthase |
| 1w7k.1.A | 15.00 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.68 | FOLC BIFUNCTIONAL PROTEIN |
| 1w78.1.A | 15.00 | monomer | HHblits | X-ray | 1.82Å | 0.27 | 0.68 | FOLC BIFUNCTIONAL PROTEIN |
| 4c12.1.A | 26.34 | homo-dimer | BLAST | X-ray | 1.80Å | 0.35 | 0.55 | UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--L-LYSINE LIGASE |
| 3lk7.1.A | 27.73 | monomer | BLAST | X-ray | 1.50Å | 0.35 | 0.50 | UDP-N-acetylmuramoylalanine--D-glutamate ligase |
| 3mvn.1.A | 27.07 | monomer | HHblits | X-ray | 1.90Å | 0.33 | 0.30 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase |
| 2obn.1.A | 15.79 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.30 | Hypothetical protein |
| 2obn.1.B | 15.79 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.30 | Hypothetical protein |
| 3zwb.1.A | 7.58 | monomer | HHblits | X-ray | 3.10Å | 0.26 | 0.30 | PEROXISOMAL BIFUNCTIONAL ENZYME |
| 3zwb.2.A | 7.58 | monomer | HHblits | X-ray | 3.10Å | 0.26 | 0.30 | PEROXISOMAL BIFUNCTIONAL ENZYME |
| 3zw8.1.B | 7.58 | homo-dimer | HHblits | X-ray | 2.50Å | 0.25 | 0.30 | PEROXISOMAL BIFUNCTIONAL ENZYME |
| 3zw8.1.A | 7.58 | homo-dimer | HHblits | X-ray | 2.50Å | 0.25 | 0.30 | PEROXISOMAL BIFUNCTIONAL ENZYME |
| 2x58.1.A | 7.63 | monomer | HHblits | X-ray | 2.80Å | 0.26 | 0.30 | PEROXISOMAL BIFUNCTIONAL ENZYME |
| 2x58.2.A | 7.63 | monomer | HHblits | X-ray | 2.80Å | 0.26 | 0.30 | PEROXISOMAL BIFUNCTIONAL ENZYME |
| 3fhl.1.A | 10.69 | homo-dimer | HHblits | X-ray | 1.93Å | 0.25 | 0.30 | Putative oxidoreductase |
| 3fhl.1.B | 10.69 | homo-dimer | HHblits | X-ray | 1.93Å | 0.25 | 0.30 | Putative oxidoreductase |
| 1u80.1.A | 11.81 | homo-dimer | HHblits | X-ray | 2.85Å | 0.27 | 0.29 | Coenzyme A biosynthesis bifunctional protein coaBC |
| 1u7u.1.A | 11.81 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.29 | Coenzyme A biosynthesis bifunctional protein coaBC |
| 1pjt.1.A | 14.17 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.29 | Siroheme synthase |
| 1pjt.1.B | 14.17 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.29 | Siroheme synthase |
| 1pjq.1.A | 15.08 | homo-dimer | HHblits | X-ray | 2.21Å | 0.26 | 0.28 | Siroheme synthase |
| 1pjq.1.B | 15.08 | homo-dimer | HHblits | X-ray | 2.21Å | 0.26 | 0.28 | Siroheme synthase |
| 1ee9.1.A | 18.03 | homo-dimer | HHblits | X-ray | 3.00Å | 0.28 | 0.27 | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE |
| 1dia.1.A | 14.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.27 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE |
| 1dia.1.B | 14.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.27 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE |
| 3u3x.1.A | 10.32 | homo-dimer | HHblits | X-ray | 2.79Å | 0.25 | 0.28 | Oxidoreductase |
| 1a4i.1.A | 14.88 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.27 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE / METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE |
| 1a4i.1.B | 14.88 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.27 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE / METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE |
| 4mkx.1.A | 11.20 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.25 | 0.28 | Inositol dehydrogenase |
| 3d4o.1.A | 16.26 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.28 | Dipicolinate synthase subunit A |
| 3d4o.1.B | 16.26 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.28 | Dipicolinate synthase subunit A |
| 5b3v.1.A | 7.81 | monomer | HHblits | X-ray | 2.59Å | 0.23 | 0.29 | Biliverdin reductase |
| 5b3u.2.A | 7.81 | monomer | HHblits | X-ray | 2.70Å | 0.23 | 0.29 | Biliverdin reductase |
| 5b3t.1.A | 7.81 | monomer | HHblits | X-ray | 2.10Å | 0.23 | 0.29 | Biliverdin reductase |
| 3moi.1.A | 11.57 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.27 | Probable dehydrogenase |
| 3ip3.1.A | 7.87 | homo-dimer | HHblits | X-ray | 2.14Å | 0.24 | 0.29 | Oxidoreductase, putative |
| 2ixa.1.A | 10.57 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.28 | ALPHA-N-ACETYLGALACTOSAMINIDASE |
| 3dfz.1.A | 11.29 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.28 | Precorrin-2 dehydrogenase |
| 3dfz.1.B | 11.29 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.28 | Precorrin-2 dehydrogenase |
| 4ply.1.A | 15.13 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.28 | 0.27 | malate dehydrogenase |
| 3do5.1.A | 14.05 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.27 | Homoserine dehydrogenase |
| 3e18.1.A | 13.01 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.28 | oxidoreductase |
| 3e18.1.B | 13.01 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.28 | oxidoreductase |
| 2yyy.1.A | 10.74 | homo-dimer | HHblits | X-ray | 1.85Å | 0.27 | 0.27 | Glyceraldehyde-3-phosphate dehydrogenase |
| 4gqa.1.A | 11.48 | homo-tetramer | HHblits | X-ray | 2.42Å | 0.25 | 0.27 | NAD binding Oxidoreductase |
| 1ofg.1.A | 11.29 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.24 | 0.28 | GLUCOSE-FRUCTOSE OXIDOREDUCTASE |
| 3evn.1.A | 10.57 | monomer | HHblits | X-ray | 2.00Å | 0.24 | 0.28 | Oxidoreductase, Gfo/Idh/MocA family |
| 3f4l.1.A | 8.87 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.28 | Putative oxidoreductase yhhX |
| 4koa.1.A | 9.02 | homo-dimer | HHblits | X-ray | 1.93Å | 0.25 | 0.27 | 1,5-anhydro-D-fructose reductase |
| 4mio.1.A | 10.66 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.25 | 0.27 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 4mio.1.D | 10.66 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.25 | 0.27 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 3q2k.1.A | 8.26 | homo-octamer | HHblits | X-ray | 2.13Å | 0.25 | 0.27 | oxidoreductase |
| 3q2k.1.B | 8.26 | homo-octamer | HHblits | X-ray | 2.13Å | 0.25 | 0.27 | oxidoreductase |
| 4had.1.A | 5.69 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.24 | 0.28 | Probable oxidoreductase protein |
| 5a02.1.A | 9.76 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.28 | ALDOSE-ALDOSE OXIDOREDUCTASE |
| 2glx.1.A | 8.20 | monomer | HHblits | X-ray | 2.20Å | 0.24 | 0.27 | 1,5-anhydro-D-fructose reductase |
| 1lld.1.A | 10.26 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.26 | L-LACTATE DEHYDROGENASE |
| 1lth.2.A | 10.26 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.26 | L-LACTATE DEHYDROGENASE (T- AND R- STATE TETRAMER COMPLEX) |
| 1evj.1.A | 10.57 | homo-dimer | HHblits | X-ray | 2.70Å | 0.23 | 0.28 | GLUCOSE-FRUCTOSE OXIDOREDUCTASE |
| 3mtj.1.A | 13.45 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.26 | 0.27 | Homoserine dehydrogenase |
| 1xea.1.A | 6.56 | homo-tetramer | HHblits | X-ray | 2.65Å | 0.24 | 0.27 | Oxidoreductase, Gfo/Idh/MocA family |
| 1zh8.1.A | 6.56 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.27 | oxidoreductase |
| 3db2.1.A | 9.09 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.27 | putative NADPH-dependent oxidoreductase |
| 3fd8.1.A | 10.74 | homo-hexamer | HHblits | X-ray | 2.45Å | 0.25 | 0.27 | Oxidoreductase, Gfo/Idh/MocA family |
| 1ryd.1.A | 8.06 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.22 | 0.28 | glucose-fructose oxidoreductase |
| 1rye.1.B | 8.06 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.22 | 0.28 | glucose-fructose oxidoreductase |
| 1rye.1.A | 8.06 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.22 | 0.28 | glucose-fructose oxidoreductase |
| 1b0a.1.A | 16.52 | homo-dimer | HHblits | X-ray | 2.56Å | 0.28 | 0.26 | PROTEIN (FOLD BIFUNCTIONAL PROTEIN) |
| 2ho5.1.A | 8.20 | homo-dimer | HHblits | X-ray | 2.56Å | 0.24 | 0.27 | Oxidoreductase, Gfo/Idh/MocA family |
| 3l07.1.A | 20.18 | homo-dimer | HHblits | X-ray | 1.88Å | 0.29 | 0.26 | Bifunctional protein folD |
| 3rc7.1.A | 9.76 | monomer | HHblits | X-ray | 2.00Å | 0.23 | 0.28 | Sugar 3-ketoreductase |
| 3rc9.1.A | 9.76 | monomer | HHblits | X-ray | 1.91Å | 0.23 | 0.28 | Sugar 3-ketoreductase |
| 3rc1.1.A | 9.76 | monomer | HHblits | X-ray | 1.71Å | 0.23 | 0.28 | Sugar 3-ketoreductase |
| 3rcb.1.A | 9.84 | monomer | HHblits | X-ray | 2.49Å | 0.23 | 0.27 | Sugar 3-ketoreductase |
| 4a5o.1.A | 17.39 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.26 | BIFUNCTIONAL PROTEIN FOLD |
| 4a5o.2.B | 17.39 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.26 | BIFUNCTIONAL PROTEIN FOLD |
| 3vtf.1.A | 15.79 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.26 | UDP-glucose 6-dehydrogenase |
| 4fb5.1.A | 9.09 | homo-tetramer | HHblits | X-ray | 2.61Å | 0.24 | 0.27 | Probable oxidoreductase protein |
| 4hkt.1.A | 7.50 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.24 | 0.27 | Inositol 2-dehydrogenase |
| 4hkt.1.B | 7.50 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.24 | 0.27 | Inositol 2-dehydrogenase |
| 3bio.1.A | 8.40 | homo-dimer | HHblits | X-ray | 1.80Å | 0.25 | 0.27 | Oxidoreductase, Gfo/Idh/MocA family |
| 3ngl.1.A | 15.93 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.25 | Bifunctional protein folD |
| 3oa2.1.A | 9.17 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.24 | 0.27 | WbpB |
| 3oa2.1.C | 9.17 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.24 | 0.27 | WbpB |
| 3kux.1.A | 10.92 | monomer | HHblits | X-ray | 2.75Å | 0.24 | 0.27 | Putative oxidoreductase |
| 3ojl.1.A | 17.12 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.25 | Cap5O |
| 3ojl.1.B | 17.12 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.25 | Cap5O |
| 3ojo.1.A | 17.12 | homo-dimer | HHblits | X-ray | 2.50Å | 0.30 | 0.25 | Cap5O |
| 3ojo.1.B | 17.12 | homo-dimer | HHblits | X-ray | 2.50Å | 0.30 | 0.25 | Cap5O |
| 2y0d.1.A | 13.16 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.27 | 0.26 | UDP-GLUCOSE DEHYDROGENASE |
| 2y0e.1.A | 13.16 | homo-dimer | HHblits | X-ray | 1.75Å | 0.26 | 0.26 | UDP-GLUCOSE DEHYDROGENASE |
| 2y0e.2.A | 13.16 | homo-dimer | HHblits | X-ray | 1.75Å | 0.26 | 0.26 | UDP-GLUCOSE DEHYDROGENASE |
| 3g0o.1.A | 12.17 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.26 | 3-hydroxyisobutyrate dehydrogenase |
| 4xrr.1.B | 15.93 | homo-dimer | HHblits | X-ray | 2.55Å | 0.27 | 0.25 | CalS8 |
| 4xrr.1.A | 15.93 | homo-dimer | HHblits | X-ray | 2.55Å | 0.27 | 0.25 | CalS8 |
| 3cky.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.25 | 2-hydroxymethyl glutarate dehydrogenase |
| 3cky.1.C | 14.29 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.25 | 2-hydroxymethyl glutarate dehydrogenase |
| 3c8m.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.25 | Homoserine dehydrogenase |
| 2c2y.1.A | 16.96 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.25 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE-METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE |
| 2c2x.1.B | 16.96 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.25 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE-METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE |
| 4qej.1.A | 11.50 | homo-hexamer | HHblits | X-ray | 2.65Å | 0.26 | 0.25 | UDP-glucose 6-dehydrogenase |
| 3prj.1.A | 11.50 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.26 | 0.25 | UDP-glucose 6-dehydrogenase |
| 4gbj.1.A | 17.27 | homo-dimer | HHblits | X-ray | 2.05Å | 0.28 | 0.25 | 6-phosphogluconate dehydrogenase NAD-binding |
| 4gbj.2.A | 17.27 | homo-dimer | HHblits | X-ray | 2.05Å | 0.28 | 0.25 | 6-phosphogluconate dehydrogenase NAD-binding |
| 2gf2.1.A | 17.27 | homo-dimer | HHblits | X-ray | 2.38Å | 0.28 | 0.25 | 3-hydroxyisobutyrate dehydrogenase |
| 3kb6.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.12Å | 0.27 | 0.25 | D-lactate dehydrogenase |
| 2nac.1.A | 14.29 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.25 | NAD-DEPENDENT FORMATE DEHYDROGENASE |
| 2nad.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.05Å | 0.27 | 0.25 | NAD-DEPENDENT FORMATE DEHYDROGENASE |
| 3fn4.1.A | 13.39 | homo-dimer | HHblits | X-ray | 1.96Å | 0.27 | 0.25 | NAD-dependent formate dehydrogenase |
| 5a9s.1.B | 15.04 | hetero-oligomer | HHblits | X-ray | 2.06Å | 0.26 | 0.25 | IMINE REDUCTASE |
| 3evt.1.A | 16.36 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.25 | Phosphoglycerate dehydrogenase |
| 5je8.1.B | 13.39 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.25 | 3-hydroxyisobutyrate dehydrogenase |
| 5je8.1.A | 13.39 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.25 | 3-hydroxyisobutyrate dehydrogenase |
| 1lc3.1.A | 10.53 | monomer | HHblits | X-ray | 1.50Å | 0.25 | 0.26 | Biliverdin Reductase A |
| 2gsd.1.A | 13.51 | homo-dimer | HHblits | X-ray | 1.95Å | 0.27 | 0.25 | NAD-dependent formate dehydrogenase |
| 5u5g.1.A | 13.39 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.25 | 6-phosphogluconate dehydrogenase |
| 1gcu.1.A | 9.57 | monomer | HHblits | X-ray | 1.40Å | 0.24 | 0.26 | BILIVERDIN REDUCTASE A |
| 2go1.1.A | 14.41 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.25 | NAD-dependent formate dehydrogenase |
| 2gug.1.A | 14.41 | homo-dimer | HHblits | X-ray | 2.28Å | 0.27 | 0.25 | Formate dehydrogenase |
| 3n7u.1.A | 13.39 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.25 | Formate dehydrogenase |
| 3jtm.1.A | 13.39 | monomer | HHblits | X-ray | 1.30Å | 0.26 | 0.25 | Formate dehydrogenase, mitochondrial |
| 5a9r.1.A | 15.18 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.25 | IMINE REDUCTASE |
| 5a9s.1.A | 15.18 | hetero-oligomer | HHblits | X-ray | 2.06Å | 0.26 | 0.25 | IMINE REDUCTASE |
| 5fwn.1.B | 15.18 | homo-dimer | HHblits | X-ray | 2.14Å | 0.26 | 0.25 | IMINE REDUCTASE |
| 1xdw.1.A | 16.82 | monomer | HHblits | X-ray | 1.98Å | 0.29 | 0.24 | NAD+-dependent (R)-2-Hydroxyglutarate Dehydrogenase |
| 3wr5.1.A | 11.61 | homo-dimer | HHblits | X-ray | 2.14Å | 0.26 | 0.25 | Formate dehydrogenase |
| 3wr5.2.B | 11.61 | homo-dimer | HHblits | X-ray | 2.14Å | 0.26 | 0.25 | Formate dehydrogenase |
| 3g79.1.A | 9.82 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.25 | NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase |
| 3g79.1.B | 9.82 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.25 | NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase |
| 3obb.1.A | 15.32 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.26 | 0.25 | Probable 3-hydroxyisobutyrate dehydrogenase |
| 2w2k.1.B | 11.61 | hetero-oligomer | HHblits | X-ray | 1.85Å | 0.26 | 0.25 | D-MANDELATE DEHYDROGENASE |
| 1gdh.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.25 | D-GLYCERATE DEHYDROGENASE |
| 2i9p.1.A | 15.32 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.26 | 0.25 | 3-hydroxyisobutyrate dehydrogenase |
| 2zyg.1.A | 16.51 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.25 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 2ekl.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.77Å | 0.28 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 4xkj.1.A | 15.74 | homo-dimer | HHblits | X-ray | 3.15Å | 0.28 | 0.24 | D-lactate dehydrogenase |
| 4xkj.1.B | 15.74 | homo-dimer | HHblits | X-ray | 3.15Å | 0.28 | 0.24 | D-lactate dehydrogenase |
| 2zya.1.A | 16.51 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.25 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 2zya.1.B | 16.51 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.25 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 4zgs.1.A | 15.45 | homo-tetramer | HHblits | X-ray | 2.46Å | 0.27 | 0.25 | Putative D-lactate dehydrogenase |
| 3w6u.1.A | 12.50 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.25 | 6-phosphogluconate dehydrogenase, NAD-binding protein |
| 2ejw.1.B | 12.50 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | Homoserine dehydrogenase |
| 2ejw.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | Homoserine dehydrogenase |
| 2ejw.2.B | 12.50 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | Homoserine dehydrogenase |
| 1vpd.1.A | 13.39 | monomer | HHblits | X-ray | 1.65Å | 0.25 | 0.25 | TARTRONATE SEMIALDEHYDE REDUCTASE |
| 4nu6.1.A | 11.82 | homo-dimer | HHblits | X-ray | 2.65Å | 0.27 | 0.25 | Phosphonate dehydrogenase |
| 2w2l.1.B | 11.71 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.26 | 0.25 | D-MANDELATE DEHYDROGENASE |
| 2w2l.1.A | 11.71 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.26 | 0.25 | D-MANDELATE DEHYDROGENASE |
| 2w2l.2.B | 11.71 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.25 | D-MANDELATE DEHYDROGENASE |
| 1sc6.1.A | 18.69 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.29 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 1sc6.1.B | 18.69 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.29 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 1sc6.1.C | 18.69 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.29 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 1sc6.1.D | 18.69 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.29 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 2dbz.1.A | 11.82 | homo-dimer | HHblits | X-ray | 2.45Å | 0.27 | 0.25 | Glyoxylate reductase |
| 1mfz.1.A | 12.61 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.25 | GDP-mannose 6-dehydrogenase |
| 4s1v.1.B | 10.81 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase-related protein |
| 4s1v.1.A | 10.81 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase-related protein |
| 4s1v.1.C | 10.81 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase-related protein |
| 3pef.1.A | 13.51 | homo-tetramer | HHblits | X-ray | 2.07Å | 0.26 | 0.25 | 6-phosphogluconate dehydrogenase, NAD-binding |
| 3pef.2.C | 13.51 | homo-tetramer | HHblits | X-ray | 2.07Å | 0.26 | 0.25 | 6-phosphogluconate dehydrogenase, NAD-binding |
| 3w6z.1.A | 14.55 | monomer | HHblits | X-ray | 1.44Å | 0.26 | 0.25 | 6-phosphogluconate dehydrogenase, NAD-binding protein |
| 4prk.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.13Å | 0.28 | 0.24 | 4-phosphoerythronate dehydrogenase |
| 1wwk.1.A | 14.81 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.24 | phosphoglycerate dehydrogenase |
| 1wwk.1.B | 14.81 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.24 | phosphoglycerate dehydrogenase |
| 3fwn.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.24 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 3fwn.1.B | 16.67 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.24 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 2wwr.1.A | 11.93 | homo-dimer | HHblits | X-ray | 2.82Å | 0.27 | 0.25 | GLYOXYLATE REDUCTASE/HYDROXYPYRUVATE REDUCTASE |
| 2wwr.1.B | 11.93 | homo-dimer | HHblits | X-ray | 2.82Å | 0.27 | 0.25 | GLYOXYLATE REDUCTASE/HYDROXYPYRUVATE REDUCTASE |
| 2gcg.1.A | 11.93 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.25 | Glyoxylate reductase/hydroxypyruvate reductase |
| 2gcg.2.B | 11.93 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.25 | Glyoxylate reductase/hydroxypyruvate reductase |
| 3naq.1.A | 12.61 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.25 | Formate dehydrogenase |
| 3l6d.1.A | 13.51 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.25 | Putative oxidoreductase |
| 3l6d.1.B | 13.51 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.25 | Putative oxidoreductase |
| 2q50.1.A | 11.93 | homo-dimer | HHblits | X-ray | 2.45Å | 0.27 | 0.25 | Glyoxylate reductase/hydroxypyruvate reductase |
| 2q50.1.B | 11.93 | homo-dimer | HHblits | X-ray | 2.45Å | 0.27 | 0.25 | Glyoxylate reductase/hydroxypyruvate reductase |
| 2q50.2.A | 11.93 | homo-dimer | HHblits | X-ray | 2.45Å | 0.27 | 0.25 | Glyoxylate reductase/hydroxypyruvate reductase |
| 2q50.2.B | 11.93 | homo-dimer | HHblits | X-ray | 2.45Å | 0.27 | 0.25 | Glyoxylate reductase/hydroxypyruvate reductase |
| 1mx3.1.A | 13.51 | monomer | HHblits | X-ray | 1.95Å | 0.25 | 0.25 | C-terminal binding protein 1 |
| 3k5p.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.28 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 2dld.1.A | 19.44 | homo-dimer | HHblits | X-ray | 2.70Å | 0.28 | 0.24 | D-LACTATE DEHYDROGENASE |
| 2dld.1.B | 19.44 | homo-dimer | HHblits | X-ray | 2.70Å | 0.28 | 0.24 | D-LACTATE DEHYDROGENASE |
| 3pdu.1.A | 13.39 | homo-tetramer | HHblits | X-ray | 1.89Å | 0.25 | 0.25 | 3-hydroxyisobutyrate dehydrogenase family protein |
| 4e5m.1.A | 11.82 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.25 | Thermostable phosphite dehydrogenase |
| 4e5m.1.B | 11.82 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.25 | Thermostable phosphite dehydrogenase |
| 1j4a.1.A | 17.43 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.25 | D-LACTATE DEHYDROGENASE |
| 4nu5.1.A | 12.84 | homo-dimer | HHblits | X-ray | 2.35Å | 0.27 | 0.25 | Phosphonate dehydrogenase |
| 4nu5.1.B | 12.84 | homo-dimer | HHblits | X-ray | 2.35Å | 0.27 | 0.25 | Phosphonate dehydrogenase |
| 4e5n.1.A | 12.84 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.25 | Thermostable phosphite dehydrogenase |
| 4e5n.3.A | 12.84 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.25 | Thermostable phosphite dehydrogenase |
| 4e5n.3.B | 12.84 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.25 | Thermostable phosphite dehydrogenase |
| 4e5n.4.A | 12.84 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.25 | Thermostable phosphite dehydrogenase |
| 3ga0.1.A | 13.64 | monomer | HHblits | X-ray | 3.40Å | 0.26 | 0.25 | C-terminal-binding protein 1 |
| 2fss.1.A | 11.71 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | formate dehydrogenase |
| 2fss.1.B | 11.71 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | formate dehydrogenase |
| 2fss.2.A | 11.71 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | formate dehydrogenase |
| 2j6i.1.A | 11.71 | homo-dimer | HHblits | X-ray | 1.55Å | 0.25 | 0.25 | FORMATE DEHYDROGENASE |
| 2j6i.2.A | 11.71 | homo-dimer | HHblits | X-ray | 1.55Å | 0.25 | 0.25 | FORMATE DEHYDROGENASE |
| 4dll.1.A | 12.61 | homo-dimer | HHblits | X-ray | 2.11Å | 0.25 | 0.25 | 2-hydroxy-3-oxopropionate reductase |
| 5dna.1.A | 11.71 | homo-dimer | HHblits | X-ray | 1.75Å | 0.25 | 0.25 | FORMATE DEHYDROGENASE |
| 5dna.2.A | 11.71 | homo-dimer | HHblits | X-ray | 1.75Å | 0.25 | 0.25 | FORMATE DEHYDROGENASE |
| 5dn9.1.A | 11.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.25 | FDH |
| 3wx0.1.A | 12.96 | homo-tetramer | HHblits | X-ray | 3.30Å | 0.27 | 0.24 | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding |
| 2w8z.1.A | 14.81 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2yq4.1.A | 14.81 | homo-tetramer | HHblits | X-ray | 3.45Å | 0.27 | 0.24 | D-ISOMER SPECIFIC 2-HYDROXYACID DEHYDROGENASE |
| 2yq4.1.C | 14.81 | homo-tetramer | HHblits | X-ray | 3.45Å | 0.27 | 0.24 | D-ISOMER SPECIFIC 2-HYDROXYACID DEHYDROGENASE |
| 4e5k.1.A | 11.93 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.25 | Phosphite dehydrogenase (thermostable variant) |
| 4e5k.2.A | 11.93 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.25 | Phosphite dehydrogenase (thermostable variant) |
| 4e5k.2.B | 11.93 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.25 | Phosphite dehydrogenase (thermostable variant) |
| 1ygy.1.A | 17.43 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase |
| 1ygy.1.B | 17.43 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase |
| 2ome.1.A | 11.71 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.25 | C-terminal-binding protein 2 |
| 2hu2.1.A | 13.64 | homo-dimer | HHblits | X-ray | 2.85Å | 0.25 | 0.25 | C-terminal-binding protein 1 |
| 1yba.1.A | 16.82 | homo-tetramer | HHblits | X-ray | 2.24Å | 0.28 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 1yba.1.C | 16.82 | homo-tetramer | HHblits | X-ray | 2.24Å | 0.28 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 1yba.1.D | 16.82 | homo-tetramer | HHblits | X-ray | 2.24Å | 0.28 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 4e5p.1.A | 11.93 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.25 | Thermostable phosphite dehydrogenase A176R variant |
| 4e5p.1.B | 11.93 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.25 | Thermostable phosphite dehydrogenase A176R variant |
| 4e5p.2.B | 11.93 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.25 | Thermostable phosphite dehydrogenase A176R variant |
| 4e5p.3.B | 11.93 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.25 | Thermostable phosphite dehydrogenase A176R variant |
| 1j49.1.A | 18.69 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.24 | D-LACTATE DEHYDROGENASE |
| 1j49.1.B | 18.69 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.24 | D-LACTATE DEHYDROGENASE |
| 3hg7.1.A | 14.68 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.25 | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| 4lcj.1.A | 12.73 | homo-dimer | HHblits | X-ray | 2.86Å | 0.25 | 0.25 | C-terminal-binding protein 2 |
| 3qha.1.A | 14.81 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.24 | Putative oxidoreductase |
| 2w90.1.A | 12.96 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2w90.1.B | 12.96 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2g76.1.A | 12.84 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase |
| 5aov.1.A | 13.76 | homo-dimer | HHblits | X-ray | 1.40Å | 0.26 | 0.25 | GLYOXYLATE REDUCTASE |
| 1pgj.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.82Å | 0.27 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE |
| 4gwg.1.A | 14.81 | monomer | HHblits | X-ray | 1.39Å | 0.27 | 0.24 | 6-phosphogluconate dehydrogenase, decarboxylating |
| 2p4q.1.A | 10.09 | homo-dimer | HHblits | X-ray | 2.37Å | 0.26 | 0.25 | 6-phosphogluconate dehydrogenase, decarboxylating 1 |
| 2iyp.1.B | 13.76 | homo-dimer | HHblits | X-ray | 2.79Å | 0.26 | 0.25 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2iyp.1.A | 13.76 | homo-dimer | HHblits | X-ray | 2.79Å | 0.26 | 0.25 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2pgd.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE |
| 4g2n.1.A | 11.93 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.26 | 0.25 | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| 4g2n.1.B | 11.93 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.26 | 0.25 | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| 4g2n.1.C | 11.93 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.26 | 0.25 | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| 3ddn.1.A | 16.51 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase |
| 3ddn.1.B | 16.51 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.26 | 0.25 | D-3-phosphoglycerate dehydrogenase |
| 2iz1.1.B | 14.81 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2iz1.1.A | 14.81 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2iyo.1.A | 13.76 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.25 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 5b37.1.A | 14.02 | homo-dimer | HHblits | X-ray | 3.40Å | 0.27 | 0.24 | Tryptophan dehydrogenase |
| 5b37.2.B | 14.02 | homo-dimer | HHblits | X-ray | 3.40Å | 0.27 | 0.24 | Tryptophan dehydrogenase |
| 2jkv.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.53Å | 0.26 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 3gg9.1.B | 10.91 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.25 | d-3-phosphoglycerate dehydrogenase oxidoreductase protein |
| 3gg9.1.A | 10.91 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.25 | d-3-phosphoglycerate dehydrogenase oxidoreductase protein |
| 3gg9.2.B | 10.91 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.25 | d-3-phosphoglycerate dehydrogenase oxidoreductase protein |
| 4d3s.1.A | 12.04 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.24 | IMINE REDUCTASE |
| 4d3s.2.B | 12.04 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.24 | IMINE REDUCTASE |
| 4d3s.4.B | 12.04 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.24 | IMINE REDUCTASE |
| 1yb4.1.A | 12.84 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.25 | 0.25 | tartronic semialdehyde reductase |
| 4cuj.1.A | 11.11 | homo-tetramer | HHblits | X-ray | 1.88Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 4cuj.1.B | 11.11 | homo-tetramer | HHblits | X-ray | 1.88Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 4cuj.1.C | 11.11 | homo-tetramer | HHblits | X-ray | 1.88Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 4cuj.1.D | 11.11 | homo-tetramer | HHblits | X-ray | 1.88Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 5uog.1.A | 13.08 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.24 | NADPH-dependent glyoxylate/hydroxypyruvate reductase |
| 5v7g.1.A | 13.08 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.24 | NADPH-dependent glyoxylate/hydroxypyruvate reductase |
| 4xyb.1.A | 6.31 | homo-dimer | HHblits | X-ray | 1.38Å | 0.24 | 0.25 | Formate dehydrogenase |
| 4xyg.1.A | 6.31 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.25 | Formate dehydrogenase |
| 5tx7.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.51Å | 0.26 | 0.24 | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| 4cuk.1.A | 11.11 | homo-tetramer | HHblits | X-ray | 2.18Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 4cuk.1.B | 11.11 | homo-tetramer | HHblits | X-ray | 2.18Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 4cuk.1.C | 11.11 | homo-tetramer | HHblits | X-ray | 2.18Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 4cuk.1.D | 11.11 | homo-tetramer | HHblits | X-ray | 2.18Å | 0.26 | 0.24 | D-LACTATE DEHYDROGENASE |
| 5g6s.1.A | 14.02 | homo-dimer | HHblits | X-ray | 2.35Å | 0.26 | 0.24 | IMINE REDUCTASE |
| 5g6r.1.A | 14.02 | homo-dimer | HHblits | X-ray | 1.82Å | 0.26 | 0.24 | IMINE REDUCTASE |
| 5ofm.1.A | 12.04 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 5n53.1.A | 11.93 | homo-dimer | HHblits | X-ray | 1.48Å | 0.25 | 0.25 | D-3-phosphoglycerate dehydrogenase |
| 3vpx.1.A | 13.21 | homo-octamer | HHblits | X-ray | 2.55Å | 0.27 | 0.24 | Leucine dehydrogenase |
| 4z0p.1.A | 11.93 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.25 | NAD-dependent dehydrogenase |
| 5vg6.1.A | 12.15 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.24 | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding |
| 4lsw.1.A | 12.04 | monomer | HHblits | X-ray | 1.64Å | 0.25 | 0.24 | D-2-hydroxyacid dehydrogensase protein |
| 4ebf.1.A | 11.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | Thermostable phosphite dehydrogenase |
| 4ebf.1.B | 11.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | Thermostable phosphite dehydrogenase |
| 4ebf.2.B | 11.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | Thermostable phosphite dehydrogenase |
| 4ebf.3.A | 11.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | Thermostable phosphite dehydrogenase |
| 2d0i.1.A | 10.19 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.24 | dehydrogenase |
| 2d0i.1.B | 10.19 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.24 | dehydrogenase |
| 5v72.1.B | 14.15 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.24 | NADPH-dependent glyoxylate/hydroxypyruvate reductase |
| 5v72.1.A | 14.15 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.24 | NADPH-dependent glyoxylate/hydroxypyruvate reductase |
| 3wwz.1.A | 10.09 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.25 | 0.25 | D-lactate dehydrogenase (Fermentative) |
| 4d3d.1.B | 12.96 | homo-dimer | HHblits | X-ray | 1.71Å | 0.25 | 0.24 | IMINE REDUCTASE |
| 4d3d.1.A | 12.96 | homo-dimer | HHblits | X-ray | 1.71Å | 0.25 | 0.24 | IMINE REDUCTASE |
| 3zhb.1.A | 11.01 | homo-dimer | HHblits | X-ray | 2.73Å | 0.25 | 0.25 | R-IMINE REDUCTASE |
| 3wnv.1.A | 13.21 | monomer | HHblits | X-ray | 1.75Å | 0.26 | 0.24 | glyoxylate reductase |
| 4weq.1.A | 13.08 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.24 | NAD-dependent dehydrogenase |
| 4hy3.1.A | 12.04 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.25 | 0.24 | phosphoglycerate oxidoreductase |
| 4hy3.1.B | 12.04 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.25 | 0.24 | phosphoglycerate oxidoreductase |
| 4hy3.1.C | 12.04 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.25 | 0.24 | phosphoglycerate oxidoreductase |
| 4hy3.1.D | 12.04 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.25 | 0.24 | phosphoglycerate oxidoreductase |
| 5unn.1.A | 13.08 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.24 | NADPH-dependent glyoxylate/hydroxypyruvate reductase |
| 1c1x.1.B | 14.02 | hetero-oligomer | HHblits | X-ray | 1.40Å | 0.26 | 0.24 | PROTEIN (L-PHENYLALANINE DEHYDROGENASE) |
| 1bxg.1.B | 14.02 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.26 | 0.24 | PHENYLALANINE DEHYDROGENASE |
| 1c1d.1.A | 14.02 | hetero-oligomer | HHblits | X-ray | 1.25Å | 0.26 | 0.24 | L-PHENYLALANINE DEHYDROGENASE |
| 1c1x.1.A | 14.02 | hetero-oligomer | HHblits | X-ray | 1.40Å | 0.26 | 0.24 | L-PHENYLALANINE DEHYDROGENASE |
| 1bw9.1.B | 14.02 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.26 | 0.24 | PHENYLALANINE DEHYDROGENASE |
| 5nzp.1.A | 12.15 | homo-dimer | HHblits | X-ray | 1.30Å | 0.25 | 0.24 | D-3-phosphoglycerate dehydrogenase |
| 1c1d.1.B | 14.02 | hetero-oligomer | HHblits | X-ray | 1.25Å | 0.25 | 0.24 | L-PHENYLALANINE DEHYDROGENASE |
| 4oqz.1.A | 9.35 | homo-dimer | HHblits | X-ray | 1.96Å | 0.25 | 0.24 | Putative oxidoreductase YfjR |
| 1bw9.1.A | 14.02 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.25 | 0.24 | PHENYLALANINE DEHYDROGENASE |
| 4zqb.1.B | 11.11 | homo-dimer | HHblits | X-ray | 1.85Å | 0.25 | 0.24 | NADP-dependent dehydrogenase |
| 4zqb.1.A | 11.11 | homo-dimer | HHblits | X-ray | 1.85Å | 0.25 | 0.24 | NADP-dependent dehydrogenase |
| 5c5i.1.B | 11.11 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.24 | NADP-dependent dehydrogenase |
| 5c5i.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.24 | NADP-dependent dehydrogenase |
| 3ba1.1.A | 12.15 | homo-dimer | HHblits | X-ray | 1.47Å | 0.25 | 0.24 | Hydroxyphenylpyruvate reductase |
| 3gvx.1.A | 10.28 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.24 | Glycerate dehydrogenase related protein |
| 3gvx.1.B | 10.28 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.24 | Glycerate dehydrogenase related protein |
| 1qp8.1.A | 8.49 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.24 | FORMATE DEHYDROGENASE |
| 1qp8.1.B | 8.49 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.24 | FORMATE DEHYDROGENASE |
| 4oqy.1.A | 9.35 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.24 | (S)-imine reductase |
| 4oqy.1.B | 9.35 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.24 | (S)-imine reductase |
| 1leh.1.A | 9.35 | homo-octamer | HHblits | X-ray | 2.20Å | 0.25 | 0.24 | LEUCINE DEHYDROGENASE |
| 1bxg.1.A | 14.15 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.25 | 0.24 | PHENYLALANINE DEHYDROGENASE |
| 4dgs.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.24 | Dehydrogenase |
| 1dxy.1.A | 12.04 | homo-dimer | HHblits | X-ray | 1.86Å | 0.24 | 0.24 | D-2-HYDROXYISOCAPROATE DEHYDROGENASE |
| 4n18.1.A | 10.28 | homo-dimer | HHblits | X-ray | 1.97Å | 0.24 | 0.24 | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
| 3kbo.1.A | 9.43 | homo-dimer | HHblits | X-ray | 2.14Å | 0.24 | 0.24 | Glyoxylate/hydroxypyruvate reductase A |
| 3pp8.1.A | 9.43 | homo-dimer | HHblits | X-ray | 2.10Å | 0.24 | 0.24 | Glyoxylate/hydroxypyruvate reductase A |
| 3kbo.2.B | 9.43 | homo-dimer | HHblits | X-ray | 2.14Å | 0.24 | 0.24 | Glyoxylate/hydroxypyruvate reductase A |
| 1wp4.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.26 | 0.23 | 3-hydroxyisobutyrate dehydrogenase |
| 1wp4.1.D | 12.50 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.26 | 0.23 | 3-hydroxyisobutyrate dehydrogenase |
| 2cuk.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.23 | glycerate dehydrogenase/glyoxylate reductase |
| 2cuk.1.B | 14.71 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.23 | glycerate dehydrogenase/glyoxylate reductase |
| 4nfy.1.A | 12.62 | homo-dimer | HHblits | X-ray | 2.45Å | 0.25 | 0.23 | D-3-phosphoglycerate dehydrogenase, putative |
| 4njm.1.A | 11.76 | homo-dimer | HHblits | X-ray | 1.79Å | 0.25 | 0.23 | D-3-phosphoglycerate dehydrogenase, putative |
| 4njo.1.B | 11.76 | homo-dimer | HHblits | X-ray | 2.22Å | 0.25 | 0.23 | D-3-phosphoglycerate dehydrogenase, putative |
| 5dzs.1.A | 16.84 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.21 | Shikimate dehydrogenase (NADP(+)) |
| 5dzs.1.B | 16.84 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.21 | Shikimate dehydrogenase (NADP(+)) |
| 4ry0.1.A | 15.46 | monomer | HHblits | X-ray | 1.40Å | 0.27 | 0.22 | Probable ribose ABC transporter, substrate-binding protein |
| 2ioy.1.A | 10.42 | monomer | HHblits | X-ray | 1.90Å | 0.25 | 0.22 | Periplasmic sugar-binding protein |
| 3ksm.1.A | 7.29 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.22 | ABC-type sugar transport system, periplasmic component |
| 1npy.1.A | 15.22 | monomer | HHblits | X-ray | 1.75Å | 0.26 | 0.21 | Hypothetical shikimate 5-dehydrogenase-like protein HI0607 |
| 4fr5.1.A | 20.22 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.20 | Shikimate dehydrogenase |
| 4fr5.2.A | 20.22 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.20 | Shikimate dehydrogenase |
| 2py6.1.A | 16.85 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.28 | 0.20 | Methyltransferase FkbM |
| 4fpx.1.A | 19.10 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.20 | Shikimate dehydrogenase |
| 4fpx.2.A | 19.10 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.20 | Shikimate dehydrogenase |
| 4foo.1.A | 17.98 | monomer | HHblits | X-ray | 2.55Å | 0.28 | 0.20 | Shikimate dehydrogenase |
| 4foo.2.A | 17.98 | monomer | HHblits | X-ray | 2.55Å | 0.28 | 0.20 | Shikimate dehydrogenase |
| 3phg.1.A | 17.98 | monomer | HHblits | X-ray | 1.57Å | 0.27 | 0.20 | Shikimate dehydrogenase |
| 3phg.2.A | 17.98 | monomer | HHblits | X-ray | 1.57Å | 0.27 | 0.20 | Shikimate dehydrogenase |
| 3phh.1.A | 17.98 | monomer | HHblits | X-ray | 1.42Å | 0.27 | 0.20 | Shikimate dehydrogenase |
| 5but.1.A | 18.39 | hetero-oligomer | HHblits | X-ray | 5.97Å | 0.29 | 0.20 | Ktr system potassium uptake protein A,Ktr system potassium uptake protein A |
| 4fsh.1.A | 17.98 | monomer | HHblits | X-ray | 2.85Å | 0.27 | 0.20 | Shikimate dehydrogenase |
| 4fsh.2.A | 17.98 | monomer | HHblits | X-ray | 2.85Å | 0.27 | 0.20 | Shikimate dehydrogenase |
| 5m6d.1.A | 13.48 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.20 | Glyceraldehyde-3-phosphate dehydrogenase |
| 4lrs.1.B | 11.24 | hetero-oligomer | HHblits | X-ray | 1.55Å | 0.27 | 0.20 | Acetaldehyde dehydrogenase |
| 4lrt.1.B | 11.24 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.27 | 0.20 | Acetaldehyde dehydrogenase |
| 1cf2.1.A | 20.93 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.19 | PROTEIN (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE) |
| 3m2t.1.A | 14.44 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.25 | 0.20 | Probable dehydrogenase |
| 1j70.1.A | 9.89 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.24 | 0.20 | ATP SULPHURYLASE |
| 1g8g.1.A | 9.89 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.24 | 0.20 | SULFATE ADENYLYLTRANSFERASE |
| 1g8f.1.A | 9.89 | homo-hexamer | HHblits | X-ray | 1.95Å | 0.24 | 0.20 | SULFATE ADENYLYLTRANSFERASE |
| 3wgi.1.B | 16.47 | homo-dimer | HHblits | X-ray | 3.25Å | 0.29 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3wg9.2.A | 16.47 | homo-dimer | HHblits | X-ray | 1.97Å | 0.29 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3wg9.2.B | 16.47 | homo-dimer | HHblits | X-ray | 1.97Å | 0.29 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3wgh.1.B | 16.47 | homo-dimer | HHblits | X-ray | 2.05Å | 0.29 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3wgg.1.A | 16.47 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3wgg.1.B | 16.47 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1nvm.1.B | 11.36 | hetero-oligomer | HHblits | X-ray | 1.70Å | 0.26 | 0.20 | acetaldehyde dehydrogenase (acylating) |
| 1nvm.2.B | 11.36 | hetero-oligomer | HHblits | X-ray | 1.70Å | 0.26 | 0.20 | acetaldehyde dehydrogenase (acylating) |
| 2hjs.1.A | 18.60 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.19 | USG-1 protein homolog |
| 3e9m.1.A | 12.36 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.20 | Oxidoreductase, Gfo/Idh/MocA family |
| 3e9m.1.B | 12.36 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.20 | Oxidoreductase, Gfo/Idh/MocA family |
| 3e9m.2.A | 12.36 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.20 | Oxidoreductase, Gfo/Idh/MocA family |
| 2vyv.1.D | 13.64 | hetero-oligomer | HHblits | X-ray | 2.38Å | 0.26 | 0.20 | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE |
| 5vz0.1.A | 13.79 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.20 | Pyruvate carboxylase |
| 5vz0.1.D | 13.79 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.20 | Pyruvate carboxylase |
| 5bnt.1.A | 17.24 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.20 | Aspartate-semialdehyde dehydrogenase |
| 5bnt.2.A | 17.24 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.20 | Aspartate-semialdehyde dehydrogenase |
| 3upl.1.A | 17.24 | homo-dimer | HHblits | X-ray | 1.50Å | 0.27 | 0.20 | Oxidoreductase |
| 3upy.1.A | 17.24 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.20 | Oxidoreductase |
| 1tlt.1.A | 11.36 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.20 | PUTATIVE OXIDOREDUCTASE (VIRULENCE FACTOR mviM HOMOLOG) |
| 1tlt.1.B | 11.36 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.20 | PUTATIVE OXIDOREDUCTASE (VIRULENCE FACTOR mviM HOMOLOG) |
| 3uuw.1.A | 12.64 | homo-tetramer | HHblits | X-ray | 1.63Å | 0.26 | 0.20 | Putative oxidoreductase with NAD(P)-binding Rossmann-fold domain |
| 4jn6.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 1.93Å | 0.25 | 0.20 | Acetaldehyde dehydrogenase |
| 5uhw.1.A | 7.78 | homo-dimer | HHblits | X-ray | 2.24Å | 0.23 | 0.20 | Oxidoreductase protein |
| 2axq.1.A | 12.64 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.20 | Saccharopine dehydrogenase |
| 5ein.1.A | 12.64 | hetero-oligomer | HHblits | X-ray | 1.70Å | 0.26 | 0.20 | N-acetyl-gamma-glutamyl-phosphate/N-acetyl-gamma-aminoadipyl-phosphate reductase |
| 3q2i.1.A | 12.64 | homo-octamer | HHblits | X-ray | 1.50Å | 0.26 | 0.20 | dehydrogenase |
| 3euw.1.A | 17.24 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.20 | Myo-inositol dehydrogenase |
| 3mvn.1.A | 41.67 | monomer | BLAST | X-ray | 1.90Å | 0.40 | 0.16 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase |
| 2vt2.1.A | 15.48 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.19 | REDOX-SENSING TRANSCRIPTIONAL REPRESSOR REX |
| 2vt2.1.B | 15.48 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.19 | REDOX-SENSING TRANSCRIPTIONAL REPRESSOR REX |
| 1brm.1.A | 16.28 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.19 | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE |
| 1gl3.1.A | 16.28 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.19 | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE |
| 3hja.1.A | 12.64 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.25 | 0.20 | Glyceraldehyde-3-phosphate dehydrogenase |
| 3hnp.1.A | 11.36 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.24 | 0.20 | Oxidoreductase |
| 3hnp.1.D | 11.36 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.24 | 0.20 | Oxidoreductase |
| 1e5q.1.A | 14.94 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.20 | SACCHAROPINE REDUCTASE |
| 1e5l.1.A | 14.94 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.20 | SACCHAROPINE REDUCTASE |
| 1e5l.1.B | 14.94 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.20 | SACCHAROPINE REDUCTASE |
| 1ff9.1.A | 14.94 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.20 | SACCHAROPINE REDUCTASE |
| 4lsm.1.B | 10.71 | homo-dimer | HHblits | X-ray | 1.65Å | 0.28 | 0.19 | Glyceraldehyde 3-phosphate dehydrogenase, cytosolic |
| 1h6c.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.24 | 0.20 | PRECURSOR FORM OF GLUCOSE-FRUCTOSE OXIDOREDUCTASE |
| 4dpk.1.A | 10.23 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.24 | 0.20 | Malonyl-CoA/succinyl-CoA reductase |
| 1tk9.1.A | 12.64 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.25 | 0.20 | Phosphoheptose isomerase 1 |
| 4zhs.1.A | 11.36 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.24 | 0.20 | Aspartate Semialdehyde Dehydrogenase |
| 4zic.1.B | 11.36 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.24 | 0.20 | Aspartate Semialdehyde Dehydrogenase |
| 4zic.1.D | 11.36 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.24 | 0.20 | Aspartate Semialdehyde Dehydrogenase |
| 4zic.2.A | 11.36 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.24 | 0.20 | Aspartate Semialdehyde Dehydrogenase |
| 4zic.2.B | 11.36 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.24 | 0.20 | Aspartate Semialdehyde Dehydrogenase |
| 3e82.1.A | 12.94 | homo-dimer | HHblits | X-ray | 2.04Å | 0.27 | 0.19 | Putative oxidoreductase |
| 3abi.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.44Å | 0.28 | 0.19 | Putative uncharacterized protein PH1688 |
| 3abi.1.B | 14.29 | homo-dimer | HHblits | X-ray | 2.44Å | 0.28 | 0.19 | Putative uncharacterized protein PH1688 |
| 5eio.1.A | 12.94 | hetero-oligomer | HHblits | X-ray | 1.80Å | 0.27 | 0.19 | N-acetyl-gamma-glutamyl-phosphate/N-acetyl-gamma-aminoadipyl-phosphate reductase |
| 2ozp.1.A | 11.76 | monomer | HHblits | X-ray | 2.01Å | 0.27 | 0.19 | N-acetyl-gamma-glutamyl-phosphate reductase |
| 5but.1.A | 16.87 | hetero-oligomer | HHblits | X-ray | 5.97Å | 0.28 | 0.19 | Ktr system potassium uptake protein A,Ktr system potassium uptake protein A |
| 3i23.1.A | 12.79 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.19 | Oxidoreductase, Gfo/Idh/MocA family |
| 5l76.1.A | 10.34 | monomer | HHblits | X-ray | 2.57Å | 0.25 | 0.20 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 5o1o.1.A | 10.34 | homo-dimer | HHblits | X-ray | 2.48Å | 0.25 | 0.20 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 5l78.1.A | 10.34 | homo-dimer | HHblits | X-ray | 2.68Å | 0.25 | 0.20 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 5l78.1.B | 10.34 | homo-dimer | HHblits | X-ray | 2.68Å | 0.25 | 0.20 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 1ydw.1.A | 8.05 | homo-dimer | HHblits | X-ray | 2.49Å | 0.25 | 0.20 | AX110P-like protein |
| 2q4e.1.B | 8.05 | homo-dimer | HHblits | X-ray | 2.49Å | 0.25 | 0.20 | Probable oxidoreductase At4g09670 |
| 4g65.1.A | 21.95 | homo-dimer | HHblits | X-ray | 2.09Å | 0.29 | 0.18 | Trk system potassium uptake protein trkA |
| 4g65.1.B | 21.95 | homo-dimer | HHblits | X-ray | 2.09Å | 0.29 | 0.18 | Trk system potassium uptake protein trkA |
| 2czc.1.A | 10.47 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.25 | 0.19 | Glyceraldehyde-3-phosphate dehydrogenase |
| 3ic5.1.A | 16.67 | monomer | HHblits | X-ray | 2.08Å | 0.27 | 0.19 | putative saccharopine dehydrogenase |
| 2gas.1.A | 11.76 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.19 | isoflavone reductase |
| 2gas.2.A | 11.76 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.19 | isoflavone reductase |
| 2yv2.1.A | 19.51 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.18 | Succinyl-CoA synthetase alpha chain |
| 3k9q.1.A | 15.48 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.19 | Glyceraldehyde-3-phosphate dehydrogenase 1 |
| 3hq4.1.D | 15.48 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.19 | Glyceraldehyde-3-phosphate dehydrogenase 1 |
| 5jya.1.A | 13.10 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.27 | 0.19 | Glyceraldehyde-3-phosphate dehydrogenase |
| 2p2s.1.A | 10.59 | homo-dimer | HHblits | X-ray | 1.25Å | 0.26 | 0.19 | Putative oxidoreductase |
| 3q11.1.A | 14.29 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 3q1l.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.19 | Aspartate beta-semialdehyde dehydrogenase |
| 3ezy.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 2.04Å | 0.26 | 0.19 | Dehydrogenase |
| 3vos.1.A | 14.12 | homo-dimer | HHblits | X-ray | 2.18Å | 0.25 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 3gdo.1.A | 9.30 | homo-dimer | HHblits | X-ray | 2.03Å | 0.24 | 0.19 | Uncharacterized oxidoreductase yvaA |
| 4gmf.1.A | 11.63 | homo-dimer | HHblits | X-ray | 1.85Å | 0.24 | 0.19 | Yersiniabactin biosynthetic protein YbtU |
| 1b7g.1.A | 10.59 | homo-dimer | HHblits | X-ray | 2.05Å | 0.25 | 0.19 | PROTEIN (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE) |
| 4gmg.1.A | 11.63 | homo-dimer | HHblits | X-ray | 2.31Å | 0.24 | 0.19 | Yersiniabactin biosynthetic protein YbtU |
| 5kvq.1.B | 11.63 | homo-dimer | HHblits | X-ray | 1.45Å | 0.24 | 0.19 | Irp3 protein |
| 1qyc.1.A | 15.48 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.19 | phenylcoumaran benzylic ether reductase PT1 |
| 1qyc.1.B | 15.48 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.19 | phenylcoumaran benzylic ether reductase PT1 |
| 3llg.1.A | 15.48 | homo-dimer | HHblits | X-ray | 2.18Å | 0.26 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 4iq0.1.A | 8.05 | homo-dimer | HHblits | X-ray | 2.00Å | 0.23 | 0.20 | Oxidoreductase, Gfo/Idh/MocA family |
| 4iq0.2.B | 8.05 | homo-dimer | HHblits | X-ray | 2.00Å | 0.23 | 0.20 | Oxidoreductase, Gfo/Idh/MocA family |
| 3tz6.1.A | 14.29 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 3wyb.1.A | 10.59 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.19 | Meso-diaminopimelate D-dehydrogenase |
| 5gz1.1.A | 10.59 | homo-dimer | HHblits | X-ray | 1.78Å | 0.25 | 0.19 | Meso-diaminopimelate D-dehydrogenase |
| 5gz3.1.A | 10.59 | homo-dimer | HHblits | X-ray | 1.59Å | 0.25 | 0.19 | Meso-diaminopimelate D-dehydrogenase |
| 5gz3.1.B | 10.59 | homo-dimer | HHblits | X-ray | 1.59Å | 0.25 | 0.19 | Meso-diaminopimelate D-dehydrogenase |
| 5gz6.1.A | 10.59 | homo-dimer | HHblits | X-ray | 1.74Å | 0.25 | 0.19 | Meso-diaminopimelate D-dehydrogenase |
| 5gz6.1.B | 10.59 | homo-dimer | HHblits | X-ray | 1.74Å | 0.25 | 0.19 | Meso-diaminopimelate D-dehydrogenase |
| 4mvj.1.A | 10.71 | monomer | HHblits | X-ray | 2.85Å | 0.25 | 0.19 | Glyceraldehyde-3-phosphate dehydrogenase A |
| 3ufx.1.A | 14.46 | hetero-oligomer | HHblits | X-ray | 2.35Å | 0.26 | 0.19 | succinyl-CoA synthetase alpha subunit |
| 3fyc.1.A | 19.75 | monomer | HHblits | X-ray | 2.15Å | 0.28 | 0.18 | Probable dimethyladenosine transferase |
| 3fyc.2.A | 19.75 | monomer | HHblits | X-ray | 2.15Å | 0.28 | 0.18 | Probable dimethyladenosine transferase |
| 3fyd.1.A | 19.75 | monomer | HHblits | X-ray | 1.75Å | 0.28 | 0.18 | Probable dimethyladenosine transferase |
| 3ksd.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.25 | 0.19 | Glyceraldehyde-3-phosphate dehydrogenase 1 |
| 4ina.1.A | 9.52 | homo-dimer | HHblits | X-ray | 2.49Å | 0.25 | 0.19 | saccharopine dehydrogenase |
| 2cdu.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.18 | NADPH OXIDASE |
| 3ikv.1.A | 13.25 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3ikv.1.B | 13.25 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3il2.1.A | 13.25 | homo-dimer | HHblits | X-ray | 2.49Å | 0.26 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3il2.1.B | 13.25 | homo-dimer | HHblits | X-ray | 2.49Å | 0.26 | 0.19 | Redox-sensing transcriptional repressor rex |
| 4f3y.1.A | 8.24 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.24 | 0.19 | Dihydrodipicolinate reductase |
| 3c1a.1.A | 11.76 | monomer | HHblits | X-ray | 1.85Å | 0.24 | 0.19 | Putative oxidoreductase |
| 4woj.1.A | 12.05 | homo-dimer | HHblits | X-ray | 2.45Å | 0.25 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 1xcb.4.A | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1xcb.1.B | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1xcb.2.A | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1xcb.2.B | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1xcb.3.A | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1xcb.3.B | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 1xcb.1.A | 13.25 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 2dt5.1.A | 13.25 | homo-dimer | HHblits | X-ray | 2.16Å | 0.25 | 0.19 | AT-rich DNA-binding protein |
| 2dt5.1.B | 13.25 | homo-dimer | HHblits | X-ray | 2.16Å | 0.25 | 0.19 | AT-rich DNA-binding protein |
| 3ikt.1.A | 13.25 | homo-dimer | HHblits | X-ray | 2.26Å | 0.25 | 0.19 | Redox-sensing transcriptional repressor rex |
| 3gfg.1.A | 9.52 | homo-dimer | HHblits | X-ray | 2.59Å | 0.25 | 0.19 | Uncharacterized oxidoreductase yvaA |
| 3o9z.1.A | 9.41 | homo-tetramer | HHblits | X-ray | 1.45Å | 0.24 | 0.19 | Lipopolysaccharide biosynthesis protein wbpB |
| 3oa0.1.A | 9.41 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.24 | 0.19 | Lipopolysaccharide biosynthesis protein wbpB |
| 1iul.1.A | 14.46 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.19 | hypothetical protein TT1466 |
| 2r00.2.A | 14.29 | homo-dimer | HHblits | X-ray | 2.03Å | 0.24 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 2r00.1.B | 14.29 | homo-dimer | HHblits | X-ray | 2.03Å | 0.24 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 2r00.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.03Å | 0.24 | 0.19 | Aspartate-semialdehyde dehydrogenase |
| 3ijp.1.A | 13.10 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.24 | 0.19 | Dihydrodipicolinate reductase |
| 3ijp.1.B | 13.10 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.24 | 0.19 | Dihydrodipicolinate reductase |
| 4dib.1.A | 10.71 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.24 | 0.19 | Glyceraldehyde 3-phosphate dehydrogenase |
| 3uw3.1.A | 14.63 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.18 | Aspartate-semialdehyde dehydrogenase |
| 3grr.1.A | 18.75 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.18 | Dimethyladenosine transferase |
| 2qw8.1.A | 13.41 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.18 | Eugenol synthase 1 |
| 4up3.1.A | 21.79 | homo-dimer | HHblits | X-ray | 1.44Å | 0.29 | 0.18 | THIOREDOXIN REDUCTASE |
| 4up3.1.B | 21.79 | homo-dimer | HHblits | X-ray | 1.44Å | 0.29 | 0.18 | THIOREDOXIN REDUCTASE |
| 5er0.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.18 | NADH oxidase |
| 3oq4.2.A | 15.00 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.18 | DBF4 |
| 3oq4.1.A | 15.00 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.18 | DBF4 |
| 3oq4.5.A | 15.00 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.18 | DBF4 |
| 3oq0.1.A | 15.00 | monomer | HHblits | X-ray | 2.70Å | 0.27 | 0.18 | DBF4 |
| 2h1r.1.A | 14.81 | monomer | HHblits | X-ray | 1.89Å | 0.26 | 0.18 | Dimethyladenosine transferase, putative |
| 3i52.1.A | 10.84 | monomer | HHblits | X-ray | 2.28Å | 0.24 | 0.19 | Putative leucoanthocyanidin reductase 1 |
| 3iwa.1.A | 16.88 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.17 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 1nhq.1.A | 16.46 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.18 | NADH PEROXIDASE |
| 1nhs.1.A | 16.46 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.18 | NADH PEROXIDASE |
| 1nhp.1.A | 16.46 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.18 | NADH PEROXIDASE |
| 1ps8.1.A | 13.58 | monomer | HHblits | X-ray | 2.40Å | 0.25 | 0.18 | Aspartate semialdehyde dehydrogenase |
| 1tb4.1.A | 13.58 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.18 | Aspartate-semialdehyde dehydrogenase |
| 3c85.1.A | 8.64 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3c85.1.B | 8.64 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3c85.2.A | 8.64 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3c85.2.B | 8.64 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3l4b.1.A | 15.00 | hetero-oligomer | HHblits | X-ray | 3.45Å | 0.26 | 0.18 | TrkA K+ Channel protien TM1088A |
| 1pqu.1.A | 13.58 | homo-dimer | HHblits | X-ray | 1.92Å | 0.25 | 0.18 | Aspartate-semialdehyde dehydrogenase |
| 3fuu.1.A | 16.46 | monomer | HHblits | X-ray | 1.53Å | 0.27 | 0.18 | Dimethyladenosine transferase |
| 3fuv.2.A | 16.46 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.18 | Dimethyladenosine transferase |
| 3fuv.3.A | 16.46 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.18 | Dimethyladenosine transferase |
| 3fuv.1.A | 16.46 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.18 | Dimethyladenosine transferase |
| 3fut.1.A | 16.46 | monomer | HHblits | X-ray | 1.52Å | 0.27 | 0.18 | Dimethyladenosine transferase |
| 3fut.2.A | 16.46 | monomer | HHblits | X-ray | 1.52Å | 0.27 | 0.18 | Dimethyladenosine transferase |
| 5l3z.1.A | 15.19 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.18 | polyketide ketoreductase SimC7 |
| 5l45.1.A | 15.19 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.18 | SimC7 |
| 5l4l.1.A | 15.19 | monomer | HHblits | X-ray | 1.20Å | 0.27 | 0.18 | SimC7 |
| 3wgo.1.B | 7.32 | homo-dimer | HHblits | X-ray | 2.05Å | 0.24 | 0.18 | Meso-diaminopimelate dehydrogenase |
| 3wgo.1.A | 7.32 | homo-dimer | HHblits | X-ray | 2.05Å | 0.24 | 0.18 | Meso-diaminopimelate dehydrogenase |
| 3wgq.1.A | 7.32 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.18 | Meso-diaminopimelate dehydrogenase |
| 3cge.1.A | 14.10 | homo-dimer | HHblits | X-ray | 2.26Å | 0.28 | 0.18 | Pyridine nucleotide-disulfide oxidoreductase, class I |
| 3cgb.1.A | 14.10 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.18 | Pyridine nucleotide-disulfide oxidoreductase, class I |
| 3oc4.1.A | 13.75 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.18 | Oxidoreductase, pyridine nucleotide-disulfide family |
| 3oc4.1.B | 13.75 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.18 | Oxidoreductase, pyridine nucleotide-disulfide family |
| 1vdc.1.A | 17.95 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.18 | NADPH DEPENDENT THIOREDOXIN REDUCTASE |
| 3r9u.1.A | 13.75 | homo-dimer | HHblits | X-ray | 2.36Å | 0.26 | 0.18 | Thioredoxin reductase |
| 4fx9.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.70Å | 0.28 | 0.18 | Coenzyme A disulfide reductase |
| 1jee.1.A | 11.25 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.26 | 0.18 | SULFATE ADENYLYLTRANSFERASE |
| 1jec.1.A | 11.25 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.18 | SULFATE ADENYLYLTRANSFERASE |
| 1f8w.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.27 | 0.18 | NADH PEROXIDASE |
| 2whd.1.A | 16.46 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.18 | THIOREDOXIN REDUCTASE |
| 2q0k.1.A | 13.92 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.18 | Thioredoxin reductase |
| 2q0k.1.B | 13.92 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.18 | Thioredoxin reductase |
| 3ish.1.B | 13.92 | homo-dimer | HHblits | X-ray | 2.43Å | 0.26 | 0.18 | Thioredoxin reductase |
| 3qa9.1.A | 16.25 | monomer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | CoA binding domain protein |
| 4xx0.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.25 | 0.18 | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial |
| 1ps9.1.A | 12.66 | monomer | HHblits | X-ray | 2.20Å | 0.26 | 0.18 | 2,4-dienoyl-CoA reductase |
| 1nhr.1.A | 15.38 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.18 | NADH PEROXIDASE |
| 3f8p.1.A | 14.10 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.18 | Thioredoxin reductase (TrxB-3) |
| 3f8r.1.B | 14.10 | homo-dimer | HHblits | X-ray | 1.95Å | 0.27 | 0.18 | Thioredoxin reductase (TrxB-3) |
| 2e6u.1.A | 17.95 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.18 | hypothetical protein PH1109 |
| 2d59.1.A | 17.95 | monomer | HHblits | X-ray | 1.65Å | 0.27 | 0.18 | hypothetical protein PH1109 |
| 3lb8.1.A | 16.88 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.17 | Putidaredoxin reductase |
| 3lb8.2.A | 16.88 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.17 | Putidaredoxin reductase |
| 1joa.1.A | 15.38 | monomer | HHblits | X-ray | 2.80Å | 0.27 | 0.18 | NADH PEROXIDASE |
| 5voh.1.A | 12.66 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.18 | NADH oxidase |
| 3q9u.1.A | 15.19 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.26 | 0.18 | CoA binding protein |
| 3fg2.1.A | 19.74 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.17 | Putative rubredoxin reductase |
| 4xyl.1.A | 13.92 | hetero-oligomer | HHblits | X-ray | 1.95Å | 0.26 | 0.18 | alpha subunit of Acyl-CoA synthetase (NDP forming) |
| 4xz3.1.A | 13.92 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.26 | 0.18 | Acyl-CoA synthetase (NDP forming) |
| 4yak.1.C | 13.92 | hetero-oligomer | HHblits | X-ray | 2.46Å | 0.26 | 0.18 | alpha subunit of Acyl-CoA synthetase (NDP forming) |
| 4ybz.1.A | 13.92 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.26 | 0.18 | alpha subunit of Acyl-CoA synthetase (NDP forming) |
| 2h63.1.A | 11.25 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.18 | Biliverdin reductase A |
| 3wb9.1.A | 6.17 | homo-hexamer | HHblits | X-ray | 1.93Å | 0.24 | 0.18 | Diaminopimelate dehydrogenase |
| 3wbb.1.A | 6.17 | homo-hexamer | HHblits | X-ray | 1.93Å | 0.24 | 0.18 | Diaminopimelate dehydrogenase |
| 3wbf.1.A | 6.17 | homo-hexamer | HHblits | X-ray | 2.12Å | 0.24 | 0.18 | Diaminopimelate dehydrogenase |
| 3klj.1.A | 15.58 | monomer | HHblits | X-ray | 2.10Å | 0.28 | 0.17 | NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) |
| 5usx.1.A | 15.38 | homo-dimer | HHblits | X-ray | 2.60Å | 0.27 | 0.18 | Thioredoxin reductase |
| 5usx.1.B | 15.38 | homo-dimer | HHblits | X-ray | 2.60Å | 0.27 | 0.18 | Thioredoxin reductase |
| 5utx.1.A | 15.38 | homo-dimer | HHblits | X-ray | 2.46Å | 0.27 | 0.18 | Thioredoxin reductase |
| 5utx.1.B | 15.38 | homo-dimer | HHblits | X-ray | 2.46Å | 0.27 | 0.18 | Thioredoxin reductase |
| 5loc.1.A | 9.88 | homo-dimer | HHblits | X-ray | 2.04Å | 0.24 | 0.18 | Meso-diaminopimelate D-dehydrogenase |
| 5loc.1.B | 9.88 | homo-dimer | HHblits | X-ray | 2.04Å | 0.24 | 0.18 | Meso-diaminopimelate D-dehydrogenase |
| 5jci.1.A | 16.67 | monomer | HHblits | X-ray | 1.70Å | 0.26 | 0.18 | Os09g0567300 protein |
| 1dap.1.A | 9.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.18 | DIAMINOPIMELIC ACID DEHYDROGENASE |
| 1dap.1.B | 9.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.18 | DIAMINOPIMELIC ACID DEHYDROGENASE |
| 2dap.1.A | 9.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.18 | DIAMINOPIMELIC ACID DEHYDROGENASE |
| 2fp4.1.A | 12.66 | hetero-oligomer | HHblits | X-ray | 2.08Å | 0.25 | 0.18 | Succinyl-CoA ligase [GDP-forming] alpha-chain, mitochondrial |
| 1y81.1.A | 15.19 | monomer | HHblits | X-ray | 1.70Å | 0.25 | 0.18 | conserved hypothetical protein |
| 3hba.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.17 | Putative phosphosugar isomerase |
| 3hba.1.B | 14.29 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.17 | Putative phosphosugar isomerase |
| 2yv1.1.A | 14.29 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.17 | Succinyl-CoA ligase [ADP-forming] subunit alpha |
| 2bc0.1.A | 16.88 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.17 | NADH Oxidase |
| 5loa.1.A | 8.64 | homo-dimer | HHblits | X-ray | 1.84Å | 0.23 | 0.18 | Meso-diaminopimelate D-dehydrogenase |
| 5loa.1.B | 8.64 | homo-dimer | HHblits | X-ray | 1.84Å | 0.23 | 0.18 | Meso-diaminopimelate D-dehydrogenase |
| 2gr3.1.A | 10.13 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.18 | ferredoxin reductase |
| 3k30.1.A | 15.19 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.18 | Histamine dehydrogenase |
| 2bc1.1.A | 16.88 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.17 | NADH Oxidase |
| 3d8x.1.A | 15.58 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.17 | Thioredoxin reductase 1 |
| 4h4p.1.A | 10.13 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.18 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 3itj.1.A | 15.58 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.17 | Thioredoxin reductase 1 |
| 3itj.1.B | 15.58 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.17 | Thioredoxin reductase 1 |
| 3itj.2.B | 15.58 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.17 | Thioredoxin reductase 1 |
| 5uth.1.A | 15.38 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.18 | Thioredoxin reductase |
| 1q1r.1.A | 17.11 | monomer | HHblits | X-ray | 1.91Å | 0.28 | 0.17 | Putidaredoxin reductase |
| 1q1w.2.A | 17.11 | monomer | HHblits | X-ray | 2.60Å | 0.28 | 0.17 | Putidaredoxin reductase |
| 4h4x.1.A | 10.13 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.18 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4fdc.1.A | 16.88 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 2a87.1.A | 12.82 | homo-dimer | HHblits | X-ray | 3.00Å | 0.26 | 0.18 | Thioredoxin reductase |
| 2a87.1.B | 12.82 | homo-dimer | HHblits | X-ray | 3.00Å | 0.26 | 0.18 | Thioredoxin reductase |
| 4z9y.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 1.63Å | 0.30 | 0.17 | 2-deoxy-D-gluconate 3-dehydrogenase |
| 1eud.1.A | 12.82 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.25 | 0.18 | SUCCINYL-COA SYNTHETASE, ALPHA CHAIN |
| 4h4z.1.A | 10.13 | homo-dimer | HHblits | X-ray | 1.95Å | 0.24 | 0.18 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 3eua.1.A | 15.79 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.17 | PUTATIVE FRUCTOSE-AMINOACID-6-PHOSPHATE DEGLYCASE |
| 5fmh.1.A | 15.58 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.17 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 2yvg.1.A | 10.26 | homo-dimer | HHblits | X-ray | 1.60Å | 0.25 | 0.18 | Ferredoxin reductase |
| 2yvj.1.A | 10.26 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Ferredoxin reductase |
| 2yvj.1.C | 10.26 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Ferredoxin reductase |
| 1d7y.1.A | 10.26 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.18 | FERREDOXIN REDUCTASE |
| 2ogu.1.A | 10.13 | homo-dimer | HHblits | X-ray | 3.23Å | 0.24 | 0.18 | Calcium-gated potassium channel mthK |
| 4nte.1.A | 14.10 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | DepH |
| 4nte.1.B | 14.10 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | DepH |
| 2rab.1.A | 18.67 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.17 | glutathione amide reductase |
| 3wsw.1.A | 17.57 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.17 | L-lactate dehydrogenase |
| 3wsv.1.B | 17.57 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.29 | 0.17 | L-lactate dehydrogenase |
| 3wsv.1.A | 17.57 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.29 | 0.17 | L-lactate dehydrogenase |
| 1xr3.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.71Å | 0.29 | 0.17 | ACTINORHODIN POLYKETIDE KETOREDUCTASE |
| 1xr3.1.B | 16.22 | homo-tetramer | HHblits | X-ray | 2.71Å | 0.29 | 0.17 | ACTINORHODIN POLYKETIDE KETOREDUCTASE |
| 1x7h.1.D | 16.22 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.17 | Putative ketoacyl reductase |
| 2xvj.1.A | 10.39 | homo-dimer | HHblits | X-ray | 2.48Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 4ntd.1.A | 11.54 | homo-dimer | HHblits | X-ray | 1.60Å | 0.25 | 0.18 | Thioredoxin reductase |
| 4jna.1.A | 14.10 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.18 | DepH |
| 4jn9.1.A | 14.10 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | DepH |
| 4jn9.1.B | 14.10 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | DepH |
| 2g0t.1.A | 17.33 | homo-dimer | HHblits | X-ray | 2.67Å | 0.28 | 0.17 | conserved hypothetical protein |
| 5jcm.1.A | 15.58 | monomer | HHblits | X-ray | 1.90Å | 0.26 | 0.17 | Os09g0567300 protein |
| 2xlt.1.A | 10.39 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xlr.1.A | 10.39 | homo-dimer | HHblits | X-ray | 2.55Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xlp.1.A | 10.39 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xve.1.A | 10.39 | homo-dimer | HHblits | X-ray | 1.99Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xvh.1.B | 10.39 | homo-dimer | HHblits | X-ray | 2.54Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 4h4t.1.A | 10.26 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.18 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 2q7v.1.A | 6.25 | homo-dimer | HHblits | X-ray | 1.90Å | 0.23 | 0.18 | Thioredoxin reductase |
| 2q7v.1.B | 6.25 | homo-dimer | HHblits | X-ray | 1.90Å | 0.23 | 0.18 | Thioredoxin reductase |
| 2xls.1.A | 10.39 | homo-dimer | HHblits | X-ray | 3.00Å | 0.26 | 0.17 | FLAVIN-CONTAINING MONOOXYGENASE |
| 4fw8.1.A | 20.55 | homo-dimer | HHblits | X-ray | 2.79Å | 0.30 | 0.16 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 4fw8.1.B | 20.55 | homo-dimer | HHblits | X-ray | 2.79Å | 0.30 | 0.16 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3fkj.1.A | 17.11 | homo-dimer | HHblits | X-ray | 2.12Å | 0.27 | 0.17 | Putative phosphosugar isomerases |
| 3ics.1.A | 10.53 | homo-dimer | HHblits | X-ray | 1.94Å | 0.27 | 0.17 | Coenzyme A-Disulfide Reductase |
| 1ldn.1.A | 17.57 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.17 | L-LACTATE DEHYDROGENASE |
| 1ldn.1.C | 17.57 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.17 | L-LACTATE DEHYDROGENASE |
| 5miu.1.A | 15.79 | monomer | HHblits | X-ray | 3.50Å | 0.27 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.A | 15.79 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.B | 15.79 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 5jcl.1.A | 14.29 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.17 | Os09g0567300 protein |
| 5m5j.1.A | 11.54 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.18 | Thioredoxin reductase |
| 5m5j.1.B | 11.54 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.18 | Thioredoxin reductase |
| 4fk1.1.A | 7.69 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.18 | Putative thioredoxin reductase |
| 5fs8.1.A | 17.11 | monomer | HHblits | X-ray | 1.40Å | 0.27 | 0.17 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs9.1.A | 17.11 | monomer | HHblits | X-ray | 1.75Å | 0.27 | 0.17 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 4h4v.1.A | 10.26 | homo-dimer | HHblits | X-ray | 1.40Å | 0.25 | 0.18 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 3gd4.1.A | 15.79 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd4.1.B | 15.79 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 1gv4.1.A | 15.79 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.17 | PROGRAMED CELL DEATH PROTEIN 8 |
| 3gd3.1.A | 15.79 | monomer | HHblits | X-ray | 2.95Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.2.A | 15.79 | monomer | HHblits | X-ray | 2.95Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.4.A | 15.79 | monomer | HHblits | X-ray | 2.95Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 4lii.1.A | 17.11 | monomer | HHblits | X-ray | 1.88Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 4bur.1.A | 17.11 | monomer | HHblits | X-ray | 2.88Å | 0.26 | 0.17 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 4bur.3.A | 17.11 | monomer | HHblits | X-ray | 2.88Å | 0.26 | 0.17 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs7.1.A | 17.11 | monomer | HHblits | X-ray | 1.85Å | 0.26 | 0.17 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 1ldb.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.28 | 0.17 | APO-L-LACTATE DEHYDROGENASE |
| 2yqu.1.A | 20.27 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.17 | 2-oxoglutarate dehydrogenase E3 component |
| 2eq7.1.A | 20.27 | hetero-oligomer | HHblits | X-ray | 1.80Å | 0.28 | 0.17 | 2-oxoglutarate dehydrogenase E3 component |
| 5x1y.1.A | 12.00 | homo-dimer | HHblits | X-ray | 3.48Å | 0.27 | 0.17 | Mercuric reductase |
| 3lls.1.A | 20.83 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.16 | 3-ketoacyl-(Acyl-carrier-protein) reductase |
| 1xhc.1.A | 17.11 | monomer | HHblits | X-ray | 2.35Å | 0.26 | 0.17 | NADH oxidase /nitrite reductase |
| 5kvi.1.A | 15.79 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 1m6i.1.A | 15.79 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.17 | Programmed cell death protein 8 |
| 2ag5.1.A | 20.83 | homo-tetramer | HHblits | X-ray | 1.84Å | 0.30 | 0.16 | dehydrogenase/reductase (SDR family) member 6 |
| 3ijr.1.A | 15.07 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.29 | 0.16 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 4h4r.1.A | 10.39 | homo-dimer | HHblits | X-ray | 1.40Å | 0.25 | 0.17 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 5iq4.1.A | 9.09 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.17 | Flavin-containing monooxygenase |
| 5gsn.1.A | 9.09 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.17 | Flavin-containing monooxygenase |
| 3ef6.1.A | 12.99 | monomer | HHblits | X-ray | 1.80Å | 0.25 | 0.17 | Toluene 1,2-dioxygenase system ferredoxin--NAD(+) reductase |
| 4emi.1.A | 12.99 | monomer | HHblits | X-ray | 1.81Å | 0.25 | 0.17 | TodA |
| 4emj.1.A | 12.99 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.25 | 0.17 | TodA |
| 3pqf.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.49Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 3pqf.1.C | 18.92 | homo-tetramer | HHblits | X-ray | 2.49Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 3pqd.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 3pqd.1.C | 18.92 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 3pqd.1.D | 18.92 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 3pqd.2.C | 18.92 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 3pqd.2.D | 18.92 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.28 | 0.17 | L-lactate dehydrogenase |
| 6b4o.1.A | 20.27 | homo-dimer | HHblits | X-ray | 1.73Å | 0.28 | 0.17 | Glutathione reductase |
| 5fs6.1.A | 17.33 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.17 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 2v3a.1.A | 15.79 | monomer | HHblits | X-ray | 2.40Å | 0.26 | 0.17 | RUBREDOXIN REDUCTASE |
| 2v3b.1.A | 15.79 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.26 | 0.17 | RUBREDOXIN REDUCTASE |
| 5ipy.1.A | 9.09 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.17 | Flavin-containing monooxygenase |
| 5kvh.1.A | 17.33 | homo-dimer | HHblits | X-ray | 2.27Å | 0.27 | 0.17 | Apoptosis-inducing factor 1, mitochondrial |
| 4amv.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.05Å | 0.27 | 0.17 | GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE [ISOMER IZING] |
| 1jxa.1.A | 13.33 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.17 | glucosamine 6-phosphate synthase |
| 1jxa.2.B | 13.33 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.17 | glucosamine 6-phosphate synthase |
| 1jxa.2.A | 13.33 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.17 | glucosamine 6-phosphate synthase |
| 2vf5.1.A | 13.33 | monomer | HHblits | X-ray | 2.90Å | 0.27 | 0.17 | GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE |
| 2j6h.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.35Å | 0.27 | 0.17 | GLUCOSAMINE-FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE |
| 3ooj.1.E | 13.33 | homo-octamer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Glucosamine/fructose-6-phosphate aminotransferase, isomerizing |
| 3ooj.1.B | 13.33 | homo-octamer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Glucosamine/fructose-6-phosphate aminotransferase, isomerizing |
| 3ooj.1.D | 13.33 | homo-octamer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Glucosamine/fructose-6-phosphate aminotransferase, isomerizing |
| 3ooj.1.A | 13.33 | homo-octamer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Glucosamine/fructose-6-phosphate aminotransferase, isomerizing |
| 3ooj.1.F | 13.33 | homo-octamer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Glucosamine/fructose-6-phosphate aminotransferase, isomerizing |
| 3ooj.1.G | 13.33 | homo-octamer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Glucosamine/fructose-6-phosphate aminotransferase, isomerizing |
| 4emw.1.A | 18.42 | homo-dimer | HHblits | X-ray | 2.39Å | 0.26 | 0.17 | Coenzyme A disulfide reductase |
| 4eqr.1.A | 18.42 | homo-dimer | HHblits | X-ray | 1.80Å | 0.25 | 0.17 | Coenzyme A disulfide reductase |
| 3qrw.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.29 | 0.16 | ketoacyl reductase |
| 3qrw.1.B | 16.44 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.29 | 0.16 | ketoacyl reductase |
| 3ri3.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 2.29Å | 0.29 | 0.16 | ketoacyl reductase |
| 3fbs.1.A | 8.97 | homo-dimer | HHblits | X-ray | 2.15Å | 0.23 | 0.18 | Oxidoreductase |
| 4em3.1.A | 18.67 | homo-dimer | HHblits | X-ray | 1.98Å | 0.26 | 0.17 | Coenzyme A disulfide reductase |
| 5u1o.1.A | 17.33 | homo-dimer | HHblits | X-ray | 2.31Å | 0.26 | 0.17 | Glutathione reductase |
| 3emk.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.28 | 0.16 | GLUCOSE/RIBITOL DEHYDROGENASE |
| 3emk.1.C | 16.44 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.28 | 0.16 | GLUCOSE/RIBITOL DEHYDROGENASE |
| 4eqw.1.A | 18.67 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.17 | Coenzyme A disulfide reductase |
| 3d5t.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.51Å | 0.27 | 0.17 | Malate dehydrogenase |
| 2tpr.1.A | 14.67 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2tpr.1.B | 14.67 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 1fea.1.A | 16.00 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 1fea.1.B | 16.00 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2g1u.1.A | 14.67 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.17 | hypothetical protein tm1088a |
| 1i01.1.D | 16.44 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.16 | BETA-KETOACYL [ACP] REDUCTASE |
| 1q7b.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.28 | 0.16 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 1i01.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.16 | BETA-KETOACYL [ACP] REDUCTASE |
| 4ywo.1.A | 10.81 | homo-dimer | HHblits | X-ray | 1.62Å | 0.27 | 0.17 | Mercuric reductase |
| 4eqs.1.A | 18.67 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.17 | Coenzyme A disulfide reductase |
| 4eqx.1.A | 18.67 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.17 | Coenzyme A disulfide reductase |
| 3q9n.1.A | 17.33 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.26 | 0.17 | CoA binding protein |
| 4kzp.1.A | 15.07 | homo-dimer | HHblits | X-ray | 1.68Å | 0.28 | 0.16 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 1ldm.1.A | 14.86 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.17 | M4 LACTATE DEHYDROGENASE |
| 4ntc.1.A | 6.49 | homo-dimer | HHblits | X-ray | 1.90Å | 0.24 | 0.17 | GliT |
| 1yqz.1.A | 18.67 | homo-dimer | HHblits | X-ray | 1.54Å | 0.26 | 0.17 | coenzyme A disulfide reductase |
| 4dna.1.A | 14.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | Probable glutathione reductase |
| 4dna.1.B | 14.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | Probable glutathione reductase |
| 1ez4.1.A | 22.54 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.16 | LACTATE DEHYDROGENASE |
| 4hp8.1.A | 14.86 | homo-tetramer | HHblits | X-ray | 1.35Å | 0.27 | 0.17 | 2-deoxy-D-gluconate 3-dehydrogenase |
| 5tr3.1.A | 17.57 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.17 | Dihydrolipoyl dehydrogenase |
| 4ln1.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.16 | L-lactate dehydrogenase 1 |
| 4lmr.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.29 | 0.16 | L-lactate dehydrogenase 1 |
| 3djg.1.A | 12.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.17 | Glutathione reductase |
| 1uls.1.A | 15.07 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.16 | putative 3-oxoacyl-acyl carrier protein reductase |
| 3vku.1.A | 23.94 | homo-tetramer | HHblits | X-ray | 1.96Å | 0.30 | 0.16 | L-lactate dehydrogenase |
| 8ldh.1.A | 14.86 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | M4 APO-LACTATE DEHYDROGENASE |
| 2gv8.1.A | 11.69 | homo-dimer | HHblits | X-ray | 2.10Å | 0.23 | 0.17 | monooxygenase |
| 2v4m.1.A | 16.00 | homo-dimer | HHblits | X-ray | 2.29Å | 0.25 | 0.17 | GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE [ISOMERIZING] 1 |
| 1tyt.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE, OXIDIZED FORM |
| 1typ.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 1aog.1.A | 14.67 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 1ges.1.A | 10.67 | homo-dimer | HHblits | X-ray | 1.74Å | 0.25 | 0.17 | GLUTATHIONE REDUCTASE |
| 4wec.1.A | 15.07 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.27 | 0.16 | Short chain dehydrogenase |
| 4new.1.A | 14.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.17 | Trypanothione reductase, putative |
| 5u4s.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.40Å | 0.28 | 0.16 | Putative short chain dehydrogenase |
| 2hmw.1.A | 18.31 | homo-octamer | HHblits | X-ray | 3.00Å | 0.29 | 0.16 | YuaA protein |
| 2hmu.1.B | 18.31 | homo-octamer | HHblits | X-ray | 2.25Å | 0.29 | 0.16 | YuaA protein |
| 2hms.1.A | 18.31 | homo-octamer | HHblits | X-ray | 2.70Å | 0.29 | 0.16 | YuaA protein |
| 1cyd.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.16 | CARBONYL REDUCTASE |
| 1xgk.1.A | 19.44 | monomer | HHblits | X-ray | 1.40Å | 0.28 | 0.16 | NITROGEN METABOLITE REPRESSION REGULATOR NMRA |
| 1e3s.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.16 | SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE |
| 2jah.1.A | 16.44 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.27 | 0.16 | CLAVULANIC ACID DEHYDROGENASE |
| 1y6j.1.A | 13.70 | monomer | HHblits | X-ray | 3.01Å | 0.27 | 0.16 | L-lactate dehydrogenase |
| 3enn.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.16 | GLUCOSE/RIBITOL DEHYDROGENASE |
| 1gxf.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.17 | TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) |
| 2qae.1.A | 13.33 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.17 | Dihydrolipoyl dehydrogenase |
| 5vps.1.A | 13.89 | monomer | HHblits | X-ray | 1.45Å | 0.28 | 0.16 | Short-chain dehydrogenase/reductase SDR |
| 2rhr.1.A | 15.28 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.28 | 0.16 | Actinorhodin Polyketide Ketoreductase |
| 2rhr.1.B | 15.28 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.28 | 0.16 | Actinorhodin Polyketide Ketoreductase |
| 2yau.1.A | 16.22 | homo-dimer | HHblits | X-ray | 3.50Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2jk6.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.95Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2x50.1.A | 16.22 | homo-dimer | HHblits | X-ray | 3.30Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 1id1.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.17 | PUTATIVE POTASSIUM CHANNEL PROTEIN |
| 5g4k.1.A | 10.81 | hetero-oligomer | HHblits | X-ray | 1.74Å | 0.26 | 0.17 | OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN |
| 1lpf.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 5u8u.1.A | 16.22 | homo-dimer | HHblits | X-ray | 1.35Å | 0.26 | 0.17 | Dihydrolipoyl dehydrogenase |
| 3lad.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.17 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 3lad.1.B | 16.22 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.17 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 3tl2.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.28 | 0.16 | Malate dehydrogenase |
| 4m1q.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.28 | 0.16 | L-lactate dehydrogenase |
| 2ewm.1.A | 9.59 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.16 | (S)-1-Phenylethanol dehydrogenase |
| 1e6w.1.A | 15.07 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.27 | 0.16 | SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE |
| 2wba.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2wpc.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2wpc.2.A | 16.22 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 2wp5.1.B | 16.22 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.17 | TRYPANOTHIONE REDUCTASE |
| 3o0h.1.A | 13.51 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.17 | Glutathione reductase |
| 5ig2.1.A | 15.28 | homo-hexamer | HHblits | X-ray | 1.80Å | 0.28 | 0.16 | Short-chain dehydrogenase/reductase SDR |
| 1vqw.1.A | 11.84 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.17 | PROTEIN WITH SIMILARITY TO FLAVIN-CONTAINING MONOOXYGENASES AND TO MAMMALIAN DIMETHYLALANINE MONOOXYGENASES |
| 4plh.1.A | 23.19 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.16 | malate dehydrogenase |
| 4plw.1.C | 23.19 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.31 | 0.16 | malate dehydrogenase |
| 5vdn.1.A | 13.33 | homo-dimer | HHblits | X-ray | 1.55Å | 0.24 | 0.17 | Glutathione oxidoreductase |
| 1geg.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.30 | 0.16 | ACETOIN REDUCTASE |
| 4m8s.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | Putative 3-oxoacyl-[acyl-carrier protein] reductase |
| 4m8s.1.B | 16.67 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | Putative 3-oxoacyl-[acyl-carrier protein] reductase |
| 4m8s.1.C | 16.67 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | Putative 3-oxoacyl-[acyl-carrier protein] reductase |
| 4wuv.1.A | 17.81 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.16 | 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase |
| 4wuv.1.B | 17.81 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.16 | 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase |
| 2aa3.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.30 | 0.16 | L-lactate dehydrogenase |
| 2aa3.1.B | 21.43 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.30 | 0.16 | L-lactate dehydrogenase |
| 2aa3.1.C | 21.43 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.30 | 0.16 | L-lactate dehydrogenase |
| 2aa3.1.D | 21.43 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.30 | 0.16 | L-lactate dehydrogenase |
| 3tjr.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.16 | short chain dehydrogenase |
| 3tjr.1.B | 12.50 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.16 | short chain dehydrogenase |
| 4xgn.1.A | 13.70 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.26 | 0.16 | 3-hydroxyacyl-CoA dehydrogenase |
| 4plf.1.A | 18.57 | homo-tetramer | HHblits | X-ray | 1.35Å | 0.30 | 0.16 | lactate dehydrogenase |
| 4plf.1.B | 18.57 | homo-tetramer | HHblits | X-ray | 1.35Å | 0.30 | 0.16 | lactate dehydrogenase |
| 2vus.1.A | 16.67 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.27 | 0.16 | NITROGEN METABOLITE REPRESSION REGULATOR NMRA |
| 1k6x.1.A | 16.67 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.16 | NmrA |
| 3gem.1.A | 17.81 | homo-tetramer | HHblits | X-ray | 1.83Å | 0.26 | 0.16 | Short chain dehydrogenase |
| 3gem.1.D | 17.81 | homo-tetramer | HHblits | X-ray | 1.83Å | 0.26 | 0.16 | Short chain dehydrogenase |
| 4j57.1.A | 10.67 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.24 | 0.17 | Thioredoxin reductase 2 |
| 1llc.1.A | 22.86 | monomer | HHblits | X-ray | 3.00Å | 0.30 | 0.16 | L-LACTATE DEHYDROGENASE |
| 1lvl.1.A | 12.33 | homo-dimer | HHblits | X-ray | 2.45Å | 0.26 | 0.16 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1bzl.1.A | 14.86 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.17 | TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) |
| 5v36.1.A | 14.86 | homo-dimer | HHblits | X-ray | 1.88Å | 0.25 | 0.17 | Glutathione reductase |
| 2zqz.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.16 | L-lactate dehydrogenase |
| 2zqy.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.29 | 0.16 | L-lactate dehydrogenase |
| 5t2u.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 5t2v.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 4q3n.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 1.97Å | 0.29 | 0.16 | MGS-M5 |
| 2q2q.1.C | 12.50 | homo-tetramer | HHblits | X-ray | 2.02Å | 0.27 | 0.16 | Beta-D-hydroxybutyrate dehydrogenase |
| 2q2q.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.02Å | 0.27 | 0.16 | Beta-D-hydroxybutyrate dehydrogenase |
| 2q2q.3.B | 12.50 | homo-dimer | HHblits | X-ray | 2.02Å | 0.27 | 0.16 | Beta-D-hydroxybutyrate dehydrogenase |
| 4b1b.1.A | 9.33 | homo-dimer | HHblits | X-ray | 2.90Å | 0.24 | 0.17 | THIOREDOXIN REDUCTASE |
| 1e3w.1.A | 16.67 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.27 | 0.16 | SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE |
| 1t2e.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.16 | L-lactate dehydrogenase |
| 2et6.1.A | 15.28 | monomer | HHblits | X-ray | 2.22Å | 0.27 | 0.16 | (3R)-hydroxyacyl-CoA dehydrogenase |
| 3grp.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.27 | 0.16 | 3-oxoacyl-(Acyl carrierprotein) reductase |
| 3grp.1.B | 12.50 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.27 | 0.16 | 3-oxoacyl-(Acyl carrierprotein) reductase |
| 4wlf.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.16 | Malate dehydrogenase, mitochondrial |
| 1t26.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.29 | 0.16 | L-lactate dehydrogenase |
| 2x8l.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.29 | 0.16 | L-LACTATE DEHYDROGENASE |
| 3uxy.1.A | 12.33 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.26 | 0.16 | Short-chain dehydrogenase/reductase SDR |
| 3uxy.1.C | 12.33 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.26 | 0.16 | Short-chain dehydrogenase/reductase SDR |
| 1get.1.A | 10.81 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.17 | GLUTATHIONE REDUCTASE |
| 1x31.1.A | 9.33 | hetero-oligomer | HHblits | X-ray | 2.15Å | 0.24 | 0.17 | Sarcosine oxidase alpha subunit |
| 4k7z.1.A | 8.00 | homo-dimer | HHblits | X-ray | 1.50Å | 0.24 | 0.17 | Mercuric reductase |
| 1nda.1.A | 13.51 | homo-dimer | HHblits | X-ray | 3.30Å | 0.25 | 0.17 | TRYPANOTHIONE OXIDOREDUCTASE |
| 1xdi.1.A | 9.46 | homo-tetramer | HHblits | X-ray | 2.81Å | 0.25 | 0.17 | Rv3303c-lpdA |
| 2z95.1.A | 23.19 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.16 | GDP-D-mannose dehydratase |
| 2z95.1.D | 23.19 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.16 | GDP-D-mannose dehydratase |
| 1vim.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 1.36Å | 0.29 | 0.16 | Hypothetical protein AF1796 |
| 1vim.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 1.36Å | 0.29 | 0.16 | Hypothetical protein AF1796 |
| 3fwz.1.A | 15.07 | homo-dimer | HHblits | X-ray | 1.79Å | 0.25 | 0.16 | Inner membrane protein ybaL |
| 1i0z.1.A | 15.49 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.16 | L-LACTATE DEHYDROGENASE H CHAIN |
| 1i0z.1.B | 15.49 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.16 | L-LACTATE DEHYDROGENASE H CHAIN |
| 1jeo.1.A | 15.49 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | HYPOTHETICAL PROTEIN MJ1247 |
| 2hjr.1.A | 23.19 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.16 | Malate dehydrogenase |
| 4plz.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.05Å | 0.28 | 0.16 | L-lactate dehydrogenase |
| 1oc4.1.A | 21.74 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.16 | L-LACTATE DEHYDROGENASE |
| 1lsu.1.A | 18.84 | monomer | HHblits | X-ray | 2.85Å | 0.29 | 0.16 | Conserved hypothetical protein yuaA |
| 5ldh.1.A | 14.08 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.27 | 0.16 | LACTATE DEHYDROGENASE |
| 4k8d.1.A | 8.11 | homo-dimer | HHblits | X-ray | 1.86Å | 0.24 | 0.17 | Mercuric reductase |
| 3fxa.1.A | 12.16 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.24 | 0.17 | SIS domain protein |
| 1cet.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.28 | 0.16 | PROTEIN (L-LACTATE DEHYDROGENASE) |
| 1ldg.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.74Å | 0.28 | 0.16 | L-LACTATE DEHYDROGENASE |
| 3ldh.1.A | 16.90 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.16 | LACTATE DEHYDROGENASE |
| 3ruc.1.A | 15.28 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.16 | WbgU |
| 5tqm.1.A | 13.89 | monomer | HHblits | X-ray | 2.90Å | 0.26 | 0.16 | Cinnamoyl-CoA Reductase |
| 5hru.1.A | 20.29 | homo-tetramer | HHblits | X-ray | 1.71Å | 0.29 | 0.16 | L-lactate dehydrogenase |
| 5hto.1.E | 20.29 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.16 | L-lactate dehydrogenase |
| 1viv.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.16 | Hypothetical protein yckF |
| 3f9i.1.A | 18.57 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.28 | 0.16 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 5k0z.1.A | 15.71 | homo-tetramer | HHblits | EM | NA | 0.28 | 0.16 | L-lactate dehydrogenase B chain |
| 2vhw.1.E | 18.57 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 2vhw.1.A | 18.57 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 2vhy.1.A | 18.57 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 2vhy.1.B | 18.57 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 3o31.1.A | 13.04 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.16 | ThermoNicotianamine Synthase |
| 3o31.1.B | 13.04 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.16 | ThermoNicotianamine Synthase |
| 4lmp.1.A | 18.57 | monomer | HHblits | X-ray | 1.95Å | 0.28 | 0.16 | Alanine dehydrogenase |
| 2voj.1.A | 18.57 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 2voe.1.A | 18.57 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 2vhv.1.A | 18.57 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.28 | 0.16 | ALANINE DEHYDROGENASE |
| 1zx9.1.A | 9.46 | homo-dimer | HHblits | X-ray | 1.90Å | 0.24 | 0.17 | Mercuric reductase |
| 4j56.1.A | 10.96 | hetero-oligomer | HHblits | X-ray | 2.37Å | 0.25 | 0.16 | Thioredoxin reductase 2 |
| 2eez.1.A | 15.49 | homo-hexamer | HHblits | X-ray | 2.71Å | 0.27 | 0.16 | Alanine dehydrogenase |
| 3fph.1.A | 13.04 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.16 | Putative uncharacterized protein |
| 3fph.1.B | 13.04 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.16 | Putative uncharacterized protein |
| 3fpj.1.B | 13.04 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.16 | Putative uncharacterized protein |
| 3fpg.1.A | 13.04 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.16 | Putative uncharacterized protein |
| 2y99.1.B | 12.33 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.24 | 0.16 | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE |
| 2y99.1.A | 12.33 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.24 | 0.16 | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE |
| 3zv6.1.A | 12.33 | homo-tetramer | HHblits | X-ray | 2.14Å | 0.24 | 0.16 | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE |
| 4k28.1.A | 15.71 | monomer | HHblits | X-ray | 2.15Å | 0.28 | 0.16 | Shikimate dehydrogenase family protein |
| 1guy.1.A | 20.29 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.16 | MALATE DEHYDROGENASE |
| 3llv.1.A | 17.39 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.16 | Exopolyphosphatase-related protein |
| 1ur5.1.A | 20.29 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.29 | 0.16 | MALATE DEHYDROGENASE |
| 1uxi.1.A | 20.29 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.16 | MALATE DEHYDROGENASE |
| 1uxj.1.B | 20.29 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.29 | 0.16 | MALATE DEHYDROGENASE |
| 3zh2.1.A | 18.57 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.16 | L-LACTATE DEHYDROGENASE |
| 3zh2.1.D | 18.57 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.16 | L-LACTATE DEHYDROGENASE |
| 4yt4.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338 |
| 4yt5.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338 |
| 4yt5.1.B | 17.65 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338 |
| 4yt8.1.B | 17.65 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338 |
| 1m3s.1.A | 18.57 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.27 | 0.16 | Hypothetical protein yckf |
| 1m3s.1.B | 18.57 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.27 | 0.16 | Hypothetical protein yckf |
| 4al4.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 1.78Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE A CHAIN |
| 5es3.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.29Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 5es3.1.B | 15.71 | homo-tetramer | HHblits | X-ray | 2.29Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 3fi9.1.A | 12.86 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | Malate dehydrogenase |
| 2ldx.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.96Å | 0.27 | 0.16 | APO-LACTATE DEHYDROGENASE |
| 2dfd.1.A | 20.29 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.16 | Malate dehydrogenase |
| 1a5z.1.A | 22.06 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.15 | L-LACTATE DEHYDROGENASE |
| 4qt0.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 3.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4ajp.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE A CHAIN |
| 4qt0.1.B | 15.71 | homo-tetramer | HHblits | X-ray | 3.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4qt0.2.A | 15.71 | homo-tetramer | HHblits | X-ray | 3.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4qt0.2.D | 15.71 | homo-tetramer | HHblits | X-ray | 3.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4okn.1.B | 15.71 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4okn.2.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4l4r.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 5kkc.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 1.86Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 5kkc.1.C | 15.71 | homo-tetramer | HHblits | X-ray | 1.86Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 1say.1.A | 21.13 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.26 | 0.16 | L-ALANINE DEHYDROGENASE |
| 1pjc.1.A | 21.13 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.26 | 0.16 | PROTEIN (L-ALANINE DEHYDROGENASE) |
| 4zvv.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4zvv.1.B | 15.71 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4zvv.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 1uxh.1.A | 18.84 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.16 | MALATE DEHYDROGENASE |
| 1uxk.1.B | 18.84 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.16 | MALATE DEHYDROGENASE |
| 1xiv.1.A | 22.06 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.29 | 0.15 | L-lactate dehydrogenase |
| 1t2f.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.16 | L-lactate dehydrogenase B chain |
| 1t2f.1.B | 14.29 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.16 | L-lactate dehydrogenase B chain |
| 2q1w.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.19Å | 0.28 | 0.16 | Putative nucleotide sugar epimerase/ dehydratase |
| 2q1w.2.A | 21.74 | homo-dimer | HHblits | X-ray | 2.19Å | 0.28 | 0.16 | Putative nucleotide sugar epimerase/ dehydratase |
| 1sov.1.A | 12.86 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | L-lactate dehydrogenase |
| 1sow.1.A | 12.86 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | L-lactate dehydrogenase |
| 5nqb.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 1.58Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 5nqb.1.C | 15.71 | homo-tetramer | HHblits | X-ray | 1.58Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4a8t.1.A | 17.39 | homo-hexamer | HHblits | X-ray | 1.59Å | 0.28 | 0.16 | PUTRESCINE CARBAMOYLTRANSFERASE |
| 2v65.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.35Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE A CHAIN |
| 4aj4.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE A CHAIN |
| 4i9h.2.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.17Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4i9h.2.C | 15.71 | homo-tetramer | HHblits | X-ray | 2.17Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4i8x.1.C | 15.71 | homo-tetramer | HHblits | X-ray | 2.23Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 3h3f.1.C | 15.71 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 3h3f.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 3h3f.2.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 3h3f.2.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4jnk.1.C | 15.71 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4jnk.1.B | 15.71 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 4m49.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 1i10.2.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE M CHAIN |
| 1i10.2.C | 15.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE M CHAIN |
| 1i10.2.B | 15.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE M CHAIN |
| 1i10.2.A | 15.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE M CHAIN |
| 1i10.1.C | 15.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE M CHAIN |
| 1i10.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-LACTATE DEHYDROGENASE M CHAIN |
| 4r68.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.11Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 5ixs.1.D | 15.71 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 1gpj.1.A | 15.71 | homo-dimer | HHblits | X-ray | 1.95Å | 0.27 | 0.16 | GLUTAMYL-TRNA REDUCTASE |
| 2d1y.1.A | 15.71 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.27 | 0.16 | hypothetical protein TT0321 |
| 4j9u.1.C | 12.86 | hetero-oligomer | HHblits | X-ray | 3.80Å | 0.27 | 0.16 | Potassium uptake protein TrkA |
| 4j9v.1.A | 12.86 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.27 | 0.16 | Potassium uptake protein TrkA |
| 4j9v.1.B | 12.86 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.27 | 0.16 | Potassium uptake protein TrkA |
| 2hsd.1.A | 12.68 | homo-tetramer | HHblits | X-ray | 2.64Å | 0.26 | 0.16 | 3-ALPHA, 20 BETA-HYDROXYSTEROID DEHYDROGENASE |
| 3d4p.1.A | 15.94 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.16 | L-lactate dehydrogenase 1 |
| 1ceq.1.A | 18.84 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.16 | PROTEIN (L-LACTATE DEHYDROGENASE) |
| 3dxf.1.A | 15.71 | monomer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | NmrA-like family domain-containing protein 1 |
| 3dxf.2.A | 15.71 | monomer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | NmrA-like family domain-containing protein 1 |
| 1v6a.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-lactate dehydrogenase A chain |
| 3fpf.1.A | 13.24 | homo-dimer | HHblits | X-ray | 1.66Å | 0.29 | 0.15 | Putative uncharacterized protein |
| 3fpf.1.B | 13.24 | homo-dimer | HHblits | X-ray | 1.66Å | 0.29 | 0.15 | Putative uncharacterized protein |
| 5tt0.1.A | 12.86 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.16 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 5tt0.1.B | 12.86 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.16 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 3p7m.1.A | 20.29 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.16 | Malate dehydrogenase |
| 4yt2.1.A | 17.91 | homo-dimer | HHblits | X-ray | 1.65Å | 0.30 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338 |
| 4yt2.1.B | 17.91 | homo-dimer | HHblits | X-ray | 1.65Å | 0.30 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ1338 |
| 6aoo.1.A | 17.39 | homo-dimer | HHblits | X-ray | 2.15Å | 0.28 | 0.16 | Malate dehydrogenase |
| 6aoo.1.B | 17.39 | homo-dimer | HHblits | X-ray | 2.15Å | 0.28 | 0.16 | Malate dehydrogenase |
| 2a94.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.29 | 0.15 | L-lactate dehydrogenase |
| 1x7d.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.60Å | 0.29 | 0.15 | ornithine cyclodeaminase |
| 2i6t.1.A | 17.39 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | UBIQUITIN-CONJUGATING ENZYME E2-LIKE ISOFORM A |
| 3dl2.1.A | 17.39 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | Ubiquitin-conjugating enzyme E2 variant 3 |
| 2i6t.1.B | 17.39 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | UBIQUITIN-CONJUGATING ENZYME E2-LIKE ISOFORM A |
| 3h3j.1.A | 14.49 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.27 | 0.16 | L-lactate dehydrogenase 1 |
| 3slg.1.A | 18.84 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | PbgP3 protein |
| 2j5q.1.A | 13.04 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.27 | 0.16 | MALATE DEHYDROGENASE |
| 9ldb.1.A | 14.49 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | LACTATE DEHYDROGENASE |
| 9ldb.1.B | 14.49 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.16 | LACTATE DEHYDROGENASE |
| 3fef.1.A | 12.68 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.25 | 0.16 | Putative glucosidase lplD, ALPHA-GALACTURONIDASE |
| 1z73.1.A | 17.39 | monomer | HHblits | X-ray | 2.50Å | 0.27 | 0.16 | protein ArnA |
| 4h7p.1.A | 18.84 | homo-dimer | HHblits | X-ray | 1.30Å | 0.27 | 0.16 | Malate dehydrogenase |
| 2fn7.1.A | 13.04 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | Lactate Dehydrogenase |
| 4nd5.2.A | 13.04 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.16 | Lactate dehydrogenase, adjacent gene encodes predicted malate dehydrogenase |
| 5xnh.1.A | 19.70 | homo-dimer | HHblits | X-ray | 1.95Å | 0.31 | 0.15 | N(4)-bis(aminopropyl)spermidine synthase |
| 1z7e.1.B | 17.39 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.27 | 0.16 | protein ArnA |
| 1z7e.1.A | 17.39 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.27 | 0.16 | protein ArnA |
| 1z7e.1.F | 17.39 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.27 | 0.16 | protein ArnA |
| 4wkg.1.D | 17.39 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.16 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.E | 17.39 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.16 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.F | 17.39 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.16 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.B | 17.39 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.16 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.C | 17.39 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.16 | Bifunctional polymyxin resistance protein ArnA |
| 3czm.1.A | 13.04 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.16 | L-lactate dehydrogenase |
| 2jjq.1.A | 22.73 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.15 | UNCHARACTERIZED RNA METHYLTRANSFERASE PYRAB10780 |
| 2vs1.1.A | 22.73 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.15 | UNCHARACTERIZED RNA METHYLTRANSFERASE PYRAB10780 |
| 4yxf.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.16 | MupS |
| 3vvc.1.A | 5.63 | homo-hexamer | HHblits | X-ray | 2.20Å | 0.25 | 0.16 | Capsular polysaccharide synthesis enzyme Cap8E |
| 4a73.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.28 | 0.15 | L-LACTATE DEHYDROGENASE |
| 4a73.1.B | 14.71 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.28 | 0.15 | L-LACTATE DEHYDROGENASE |
| 4a73.1.C | 14.71 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.28 | 0.15 | L-LACTATE DEHYDROGENASE |
| 4a73.1.D | 14.71 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.28 | 0.15 | L-LACTATE DEHYDROGENASE |
| 1z7b.1.A | 17.39 | monomer | HHblits | X-ray | 2.31Å | 0.27 | 0.16 | protein ArnA |
| 1obb.1.A | 15.94 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | ALPHA-GLUCOSIDASE |
| 1obb.1.B | 15.94 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.16 | ALPHA-GLUCOSIDASE |
| 5f5l.1.A | 14.49 | homo-dimer | HHblits | X-ray | 1.68Å | 0.27 | 0.16 | Monooxygenase |
| 4bgv.1.A | 14.49 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.27 | 0.16 | MALATE DEHYDROGENASE |
| 4bgv.1.C | 14.49 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.27 | 0.16 | MALATE DEHYDROGENASE |
| 1u9j.1.A | 15.94 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.16 | Hypothetical protein yfbG |
| 4n7r.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.26 | 0.16 | Glutamyl-tRNA reductase 1, chloroplastic |
| 4n7r.1.C | 14.29 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.26 | 0.16 | Glutamyl-tRNA reductase 1, chloroplastic |
| 5che.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.26 | 0.16 | Glutamyl-tRNA reductase 1, chloroplastic |
| 5che.1.B | 14.29 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.26 | 0.16 | Glutamyl-tRNA reductase 1, chloroplastic |
| 3w1v.1.B | 5.63 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.25 | 0.16 | Capsular polysaccharide synthesis enzyme Cap8E |
| 3vvb.1.F | 5.63 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.25 | 0.16 | CapE |
| 5lnk.1.R | 18.84 | hetero-oligomer | HHblits | EM | 3.90Å | 0.27 | 0.16 | Mitochondrial complex I, 39 kDa subunit |
| 2x0r.1.A | 11.59 | homo-tetramer | HHblits | X-ray | 2.92Å | 0.27 | 0.16 | MALATE DEHYDROGENASE |
| 2b69.1.A | 15.28 | homo-dimer | HHblits | X-ray | 1.21Å | 0.24 | 0.16 | UDP-glucuronate decarboxylase 1 |
| 4lvc.1.A | 13.24 | homo-tetramer | HHblits | X-ray | 1.74Å | 0.28 | 0.15 | S-adenosyl-L-homocysteine hydrolase (SAHase) |
| 4lvc.1.D | 13.24 | homo-tetramer | HHblits | X-ray | 1.74Å | 0.28 | 0.15 | S-adenosyl-L-homocysteine hydrolase (SAHase) |
| 5m5k.1.D | 13.24 | homo-tetramer | HHblits | X-ray | 1.84Å | 0.28 | 0.15 | Adenosylhomocysteinase |
| 5f5n.1.A | 14.49 | homo-dimer | HHblits | X-ray | 1.30Å | 0.27 | 0.16 | Monooxygenase |
| 2d4a.1.A | 14.49 | homo-tetramer | HHblits | X-ray | 2.87Å | 0.27 | 0.16 | Malate dehydrogenase |
| 4gll.1.A | 15.28 | homo-dimer | HHblits | X-ray | 2.50Å | 0.23 | 0.16 | UDP-glucuronic acid decarboxylase 1 |
| 4gll.1.B | 15.28 | homo-dimer | HHblits | X-ray | 2.50Å | 0.23 | 0.16 | UDP-glucuronic acid decarboxylase 1 |
| 2wmd.1.A | 15.94 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.16 | NMRA-LIKE FAMILY DOMAIN CONTAINING PROTEIN 1 |
| 2wm3.1.A | 15.94 | monomer | HHblits | X-ray | 1.85Å | 0.27 | 0.16 | NMRA-LIKE FAMILY DOMAIN CONTAINING PROTEIN 1 |
| 4j2o.1.A | 10.00 | homo-hexamer | HHblits | X-ray | 2.65Å | 0.26 | 0.16 | UDP-N-acetylglucosamine 4,6-dehydratase/5-epimerase |
| 4j2o.1.D | 10.00 | homo-hexamer | HHblits | X-ray | 2.65Å | 0.26 | 0.16 | UDP-N-acetylglucosamine 4,6-dehydratase/5-epimerase |
| 4xdb.1.A | 11.27 | homo-dimer | HHblits | X-ray | 3.32Å | 0.24 | 0.16 | NADH dehydrogenase-like protein SAOUHSC_00878 |
| 2exx.1.A | 15.94 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.16 | HSCARG protein |
| 2exx.1.B | 15.94 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.16 | HSCARG protein |
| 2x0i.1.B | 15.94 | homo-dimer | HHblits | X-ray | 2.91Å | 0.26 | 0.16 | MALATE DEHYDROGENASE |
| 2x0j.1.A | 15.94 | homo-dimer | HHblits | X-ray | 2.79Å | 0.26 | 0.16 | MALATE DEHYDROGENASE |
| 4ror.1.A | 15.94 | monomer | HHblits | X-ray | 1.66Å | 0.26 | 0.16 | Malate dehydrogenase |
| 4ros.1.A | 15.94 | monomer | HHblits | X-ray | 1.95Å | 0.26 | 0.16 | Malate dehydrogenase |
| 4a7p.1.A | 11.76 | homo-dimer | HHblits | X-ray | 3.40Å | 0.28 | 0.15 | UDP-GLUCOSE DEHYDROGENASE |
| 1hyh.1.A | 16.18 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.15 | L-2-HYDROXYISOCAPROATE DEHYDROGENASE |
| 1hyh.1.C | 16.18 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.15 | L-2-HYDROXYISOCAPROATE DEHYDROGENASE |
| 5xtb.1.I | 14.29 | hetero-oligomer | HHblits | EM | NA | 0.25 | 0.16 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial |
| 3x2e.1.A | 17.91 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 3x2e.1.B | 17.91 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 3x2f.1.A | 17.91 | homo-tetramer | HHblits | X-ray | 2.04Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 3x2f.1.B | 17.91 | homo-tetramer | HHblits | X-ray | 2.04Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 3gvi.1.A | 15.94 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.16 | Malate dehydrogenase |
| 3ond.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 1.17Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 3ond.1.B | 16.42 | homo-tetramer | HHblits | X-ray | 1.17Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 2izz.1.A | 18.18 | homo-10-mer | HHblits | X-ray | 1.95Å | 0.30 | 0.15 | PYRROLINE-5-CARBOXYLATE REDUCTASE 1 |
| 3e5m.1.A | 15.94 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.16 | NmrA-like family domain-containing protein 1 |
| 3e5m.2.A | 15.94 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.16 | NmrA-like family domain-containing protein 1 |
| 3om9.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 1.98Å | 0.28 | 0.15 | Lactate dehydrogenase |
| 3l05.1.A | 13.43 | homo-trimer | HHblits | X-ray | 2.80Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 3m4n.1.A | 13.43 | homo-trimer | HHblits | X-ray | 1.90Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 4j7c.1.A | 18.18 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.15 | Ktr system potassium uptake protein A |
| 3gg2.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.27 | 0.15 | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family |
| 3p19.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.15 | Putative blue fluorescent protein |
| 1gv0.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.15 | MALATE DEHYDROGENASE |
| 1pzg.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.28 | 0.15 | lactate dehydrogenase |
| 1pze.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.28 | 0.15 | lactate dehydrogenase |
| 4avy.1.A | 10.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.16 | PROBABLE SHORT-CHAIN DEHYDROGENASE |
| 3ce6.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.27 | 0.15 | Adenosylhomocysteinase |
| 1d3a.1.D | 11.76 | homo-tetramer | HHblits | X-ray | 2.94Å | 0.27 | 0.15 | HALOPHILIC MALATE DEHYDROGENASE |
| 1hlp.1.A | 11.76 | homo-dimer | HHblits | X-ray | 3.20Å | 0.27 | 0.15 | MALATE DEHYDROGENASE |
| 1guz.1.A | 13.24 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.15 | MALATE DEHYDROGENASE |
| 3gd5.1.A | 10.14 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.26 | 0.16 | Ornithine carbamoyltransferase |
| 3geg.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.15 | Short-chain dehydrogenase/reductase SDR |
| 2hlp.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 2.59Å | 0.27 | 0.15 | MALATE DEHYDROGENASE |
| 1o6z.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.27 | 0.15 | MALATE DEHYDROGENASE |
| 2pzj.1.A | 14.08 | homo-dimer | HHblits | X-ray | 1.90Å | 0.24 | 0.16 | Putative nucleotide sugar epimerase/ dehydratase |
| 5idw.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.15 | Short-chain dehydrogenase/reductase SDR |
| 1fb5.1.A | 11.76 | homo-trimer | HHblits | X-ray | 3.50Å | 0.27 | 0.15 | ORNITHINE TRANSCARBAMOYLASE |
| 1ib6.1.A | 19.40 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.15 | MALATE DEHYDROGENASE |
| 1ie3.1.A | 19.40 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.15 | MALATE DEHYDROGENASE |
| 3l04.1.A | 11.94 | homo-trimer | HHblits | X-ray | 2.50Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 3l06.1.A | 11.94 | homo-trimer | HHblits | X-ray | 2.81Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 3m5d.1.A | 11.94 | homo-trimer | HHblits | X-ray | 2.20Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 3kzc.1.A | 11.94 | homo-trimer | HHblits | X-ray | 2.20Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 3m5c.1.A | 11.94 | homo-trimer | HHblits | X-ray | 1.85Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 3l02.1.A | 11.94 | homo-trimer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | N-acetylornithine carbamoyltransferase |
| 1hyg.1.A | 13.24 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.27 | 0.15 | L-LACTATE/MALATE DEHYDROGENASE |
| 1hye.1.A | 13.24 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.15 | L-LACTATE/MALATE DEHYDROGENASE |
| 4nwz.1.A | 7.04 | homo-dimer | HHblits | X-ray | 2.50Å | 0.23 | 0.16 | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| 3ccf.1.A | 16.67 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.15 | Cyclopropane-fatty-acyl-phospholipid synthase |
| 3ccf.2.A | 16.67 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.15 | Cyclopropane-fatty-acyl-phospholipid synthase |
| 3vtf.1.A | 14.49 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.16 | UDP-glucose 6-dehydrogenase |
| 1o5i.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.28 | 0.15 | 3-oxoacyl-(acyl carrier protein) reductase |
| 4lk3.1.A | 14.08 | homo-hexamer | HHblits | X-ray | 2.64Å | 0.23 | 0.16 | UDP-glucuronic acid decarboxylase 1 |
| 3u95.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.15 | Glycoside hydrolase, family 4 |
| 1s6y.1.A | 17.39 | monomer | HHblits | X-ray | 2.31Å | 0.25 | 0.16 | 6-phospho-beta-glucosidase |
| 5tov.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 5tov.1.B | 18.18 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.15 | Adenosylhomocysteinase |
| 2bka.1.A | 20.90 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.15 | TAT-INTERACTING PROTEIN TIP30 |
| 4bgu.1.A | 10.29 | homo-tetramer | HHblits | X-ray | 1.49Å | 0.26 | 0.15 | MALATE DEHYDROGENASE |
| 3oj0.1.A | 14.49 | monomer | HHblits | X-ray | 1.65Å | 0.25 | 0.16 | Glutamyl-tRNA reductase |
| 2v6b.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.26 | 0.15 | L-LACTATE DEHYDROGENASE |
| 4xeh.1.A | 23.08 | monomer | HHblits | X-ray | 1.39Å | 0.30 | 0.15 | Ketol-acid reductoisomerase |
| 4xdz.1.A | 23.08 | homo-dimer | HHblits | X-ray | 1.15Å | 0.30 | 0.15 | Ketol-acid reductoisomerase |
| 5v96.1.A | 17.91 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.15 | S-adenosyl-l-homocysteine Hydrolase |
| 3dhy.1.A | 11.94 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.15 | Adenosylhomocysteinase |
| 3n58.1.A | 11.94 | homo-tetramer | HHblits | X-ray | 2.39Å | 0.27 | 0.15 | Adenosylhomocysteinase |
| 3n58.1.B | 11.94 | homo-tetramer | HHblits | X-ray | 2.39Å | 0.27 | 0.15 | Adenosylhomocysteinase |
| 4nf2.1.A | 16.18 | homo-trimer | HHblits | X-ray | 1.74Å | 0.26 | 0.15 | Ornithine carbamoyltransferase |
| 4ep1.1.A | 16.18 | homo-trimer | HHblits | X-ray | 3.25Å | 0.26 | 0.15 | Ornithine carbamoyltransferase |
| 4oh7.1.A | 11.59 | homo-trimer | HHblits | X-ray | 1.50Å | 0.25 | 0.16 | Ornithine carbamoyltransferase |
| 3ulk.2.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | Ketol-acid reductoisomerase |
| 1yrl.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.15 | Ketol-acid reductoisomerase |
| 5uax.1.A | 18.46 | homo-10-mer | HHblits | X-ray | 1.85Å | 0.30 | 0.15 | Pyrroline-5-carboxylate reductase 1, mitochondrial |
| 2gra.3.A | 18.46 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.15 | Pyrroline-5-carboxylate reductase 1 |
| 2gra.2.A | 18.46 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.15 | Pyrroline-5-carboxylate reductase 1 |
| 2gra.1.A | 18.46 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.15 | Pyrroline-5-carboxylate reductase 1 |
| 2gra.4.A | 18.46 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.15 | Pyrroline-5-carboxylate reductase 1 |
| 2gra.5.A | 18.46 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.15 | Pyrroline-5-carboxylate reductase 1 |
| 3t38.1.A | 14.93 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.15 | Arsenate Reductase |
| 3t38.1.B | 14.93 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.15 | Arsenate Reductase |
| 3mtg.1.A | 13.24 | homo-tetramer | HHblits | X-ray | 2.64Å | 0.26 | 0.15 | Putative adenosylhomocysteinase 2 |
| 4j90.1.A | 18.46 | homo-octamer | HHblits | X-ray | 3.24Å | 0.29 | 0.15 | Ktr system potassium uptake protein A |
| 4j91.1.A | 18.46 | homo-octamer | HHblits | X-ray | 2.93Å | 0.29 | 0.15 | Ktr system potassium uptake protein A |
| 4j91.1.B | 18.46 | homo-octamer | HHblits | X-ray | 2.93Å | 0.29 | 0.15 | Ktr system potassium uptake protein A |
| 4j91.1.C | 18.46 | homo-octamer | HHblits | X-ray | 2.93Å | 0.29 | 0.15 | Ktr system potassium uptake protein A |
| 5k2m.1.A | 5.48 | hetero-oligomer | HHblits | X-ray | 2.18Å | 0.20 | 0.16 | RimK-related lysine biosynthesis protein |
| 1v8b.1.A | 14.93 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.15 | adenosylhomocysteinase |
| 1lss.1.A | 19.70 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | Trk system potassium uptake protein trkA homolog |
| 1lss.1.B | 19.70 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | Trk system potassium uptake protein trkA homolog |
| 1lss.2.A | 19.70 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | Trk system potassium uptake protein trkA homolog |
| 1lss.2.B | 19.70 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | Trk system potassium uptake protein trkA homolog |
| 3h9u.1.A | 14.93 | homo-octamer | HHblits | X-ray | 1.90Å | 0.27 | 0.15 | Adenosylhomocysteinase |
| 2hk8.1.A | 14.93 | monomer | HHblits | X-ray | 2.35Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 2hk8.3.A | 14.93 | monomer | HHblits | X-ray | 2.35Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 2hk8.4.A | 14.93 | monomer | HHblits | X-ray | 2.35Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 2hk8.5.A | 14.93 | monomer | HHblits | X-ray | 2.35Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 2hk8.7.A | 14.93 | monomer | HHblits | X-ray | 2.35Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 2hk7.1.A | 14.93 | monomer | HHblits | X-ray | 2.50Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 2hk7.2.A | 14.93 | monomer | HHblits | X-ray | 2.50Å | 0.27 | 0.15 | Shikimate dehydrogenase |
| 5gup.12.A | 16.18 | monomer | HHblits | EM | NA | 0.26 | 0.15 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial |
| 4tqg.1.A | 11.59 | monomer | HHblits | X-ray | 2.20Å | 0.25 | 0.16 | Putative dTDP-d-glucose 4 6-dehydratase |
| 4yte.1.A | 15.38 | monomer | HHblits | X-ray | 2.15Å | 0.29 | 0.15 | H(2)-forming methylenetetrahydromethanopterin dehydrogenase-related protein MJ0715 |
| 4j9u.1.C | 23.08 | hetero-oligomer | HHblits | X-ray | 3.80Å | 0.29 | 0.15 | Potassium uptake protein TrkA |
| 4j9v.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.29 | 0.15 | Potassium uptake protein TrkA |
| 4j9v.1.B | 23.08 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.29 | 0.15 | Potassium uptake protein TrkA |
| 2rir.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.28 | 0.15 | Dipicolinate synthase, A chain |
| 2rir.1.B | 15.15 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.28 | 0.15 | Dipicolinate synthase, A chain |
| 3r6d.1.A | 10.45 | monomer | HHblits | X-ray | 1.25Å | 0.27 | 0.15 | NAD-dependent epimerase/dehydratase |
| 4hnh.1.A | 10.45 | monomer | HHblits | X-ray | 1.58Å | 0.27 | 0.15 | NAD-dependent epimerase/dehydratase |
| 2fmu.1.A | 17.91 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.15 | HIV-1 tat interactive protein 2, 30 kDa homolog |
| 5e4r.1.A | 23.44 | monomer | HHblits | X-ray | 1.94Å | 0.30 | 0.14 | Ketol-acid reductoisomerase |
| 5hm8.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 5jxw.1.D | 16.42 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 5jpi.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 5tj9.1.C | 16.42 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 3gt0.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.14 | Pyrroline-5-carboxylate reductase |
| 4m55.1.A | 14.29 | homo-hexamer | HHblits | X-ray | 2.86Å | 0.23 | 0.16 | UDP-glucuronic acid decarboxylase 1 |
| 4m55.1.D | 14.29 | homo-hexamer | HHblits | X-ray | 2.86Å | 0.23 | 0.16 | UDP-glucuronic acid decarboxylase 1 |
| 3glq.1.A | 11.94 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 3d64.1.A | 11.94 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 3iv6.1.A | 14.93 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.15 | Putative Zn-dependent Alcohol Dehydrogenase |
| 3iv6.2.A | 14.93 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.15 | Putative Zn-dependent Alcohol Dehydrogenase |
| 3c3x.1.A | 11.59 | monomer | HHblits | X-ray | 2.15Å | 0.24 | 0.16 | Eugenol synthase 1 |
| 4f2g.1.A | 10.14 | homo-dimer | HHblits | X-ray | 2.10Å | 0.24 | 0.16 | Ornithine carbamoyltransferase 1 |
| 4kqw.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.39Å | 0.27 | 0.15 | Ketol-acid reductoisomerase |
| 3gvp.1.A | 13.43 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.15 | Adenosylhomocysteinase 3 |
| 3gvp.1.C | 13.43 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.15 | Adenosylhomocysteinase 3 |
| 3gvp.1.D | 13.43 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.15 | Adenosylhomocysteinase 3 |
| 4xdy.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.53Å | 0.27 | 0.15 | Ketol-acid reductoisomerase |
| 2c59.1.A | 16.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.15 | GDP-MANNOSE-3', 5'-EPIMERASE |
| 3ius.1.A | 11.76 | monomer | HHblits | X-ray | 1.66Å | 0.25 | 0.15 | uncharacterized conserved protein |
| 3ius.2.A | 11.76 | monomer | HHblits | X-ray | 1.66Å | 0.25 | 0.15 | uncharacterized conserved protein |
| 5c3m.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 3.06Å | 0.25 | 0.15 | Putative 6-phospho-beta-glucosidase |
| 4kqx.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.15 | Ketol-acid reductoisomerase |
| 5tqv.1.A | 11.94 | homo-dimer | HHblits | X-ray | 1.65Å | 0.26 | 0.15 | NADPH-dependent carbonyl reductase |
| 5u1p.1.A | 11.94 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.15 | NADPH-dependent carbonyl reductase |
| 4ipt.1.A | 10.45 | monomer | HHblits | X-ray | 1.55Å | 0.26 | 0.15 | NAD-dependent epimerase/dehydratase |
| 1qyd.1.A | 10.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.16 | pinoresinol-lariciresinol reductase |
| 1qyd.1.B | 10.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.16 | pinoresinol-lariciresinol reductase |
| 1qyd.2.A | 10.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.16 | pinoresinol-lariciresinol reductase |
| 1qyd.2.B | 10.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.16 | pinoresinol-lariciresinol reductase |
| 2pzk.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.15 | Putative nucleotide sugar epimerase/ dehydratase |
| 3g1u.1.A | 11.94 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 2r6j.1.A | 13.24 | monomer | HHblits | X-ray | 1.50Å | 0.25 | 0.15 | Eugenol synthase 1 |
| 4xrr.1.B | 8.82 | homo-dimer | HHblits | X-ray | 2.55Å | 0.25 | 0.15 | CalS8 |
| 4xrr.1.A | 8.82 | homo-dimer | HHblits | X-ray | 2.55Å | 0.25 | 0.15 | CalS8 |
| 4ypo.1.A | 15.38 | homo-dimer | HHblits | X-ray | 1.00Å | 0.28 | 0.15 | Ketol-acid reductoisomerase |
| 4ypo.1.B | 15.38 | homo-dimer | HHblits | X-ray | 1.00Å | 0.28 | 0.15 | Ketol-acid reductoisomerase |
| 1u8x.1.A | 11.59 | monomer | HHblits | X-ray | 2.05Å | 0.23 | 0.16 | Maltose-6'-phosphate glucosidase |
| 3ew7.1.A | 13.64 | monomer | HHblits | X-ray | 2.73Å | 0.27 | 0.15 | Lmo0794 protein |
| 2ef0.1.A | 18.18 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.15 | Ornithine carbamoyltransferase |
| 2c5e.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.15 | GDP-MANNOSE-3', 5'-EPIMERASE |
| 5uav.4.B | 17.19 | homo-dimer | HHblits | X-ray | 1.85Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase 1, mitochondrial |
| 2ger.4.A | 17.19 | monomer | HHblits | X-ray | 3.10Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase 1 |
| 2ger.2.A | 17.19 | monomer | HHblits | X-ray | 3.10Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase 1 |
| 2ger.3.A | 17.19 | monomer | HHblits | X-ray | 3.10Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase 1 |
| 2ger.1.A | 17.19 | monomer | HHblits | X-ray | 3.10Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase 1 |
| 2ger.5.A | 17.19 | monomer | HHblits | X-ray | 3.10Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase 1 |
| 6aph.1.A | 14.93 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.26 | 0.15 | Adenosylhomocysteinase |
| 1ml4.1.A | 11.76 | homo-trimer | HHblits | X-ray | 1.80Å | 0.24 | 0.15 | Aspartate Transcarbamoylase |
| 1z82.1.A | 17.19 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.14 | glycerol-3-phosphate dehydrogenase |
| 2c5a.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.40Å | 0.24 | 0.15 | GDP-MANNOSE-3', 5'-EPIMERASE |
| 3dhn.1.A | 15.15 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.15 | NAD-dependent epimerase/dehydratase |
| 2r2g.1.A | 11.76 | homo-dimer | HHblits | X-ray | 1.80Å | 0.24 | 0.15 | Eugenol synthase 1 |
| 1up7.1.A | 16.18 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.24 | 0.15 | 6-PHOSPHO-BETA-GLUCOSIDASE |
| 1up6.1.A | 16.18 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.24 | 0.15 | 6-PHOSPHO-BETA-GLUCOSIDASE |
| 3fr8.1.A | 13.43 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.15 | Putative ketol-acid reductoisomerase (Os05g0573700 protein) |
| 3fr8.1.B | 13.43 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.15 | Putative ketol-acid reductoisomerase (Os05g0573700 protein) |
| 3fr7.1.A | 13.43 | homo-dimer | HHblits | X-ray | 1.55Å | 0.25 | 0.15 | Putative ketol-acid reductoisomerase (Os05g0573700 protein) |
| 3fr7.1.B | 13.43 | homo-dimer | HHblits | X-ray | 1.55Å | 0.25 | 0.15 | Putative ketol-acid reductoisomerase (Os05g0573700 protein) |
| 5w49.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.15 | Adenosylhomocysteinase |
| 3dzb.1.A | 18.46 | homo-dimer | HHblits | X-ray | 2.46Å | 0.27 | 0.15 | Prephenate dehydrogenase |
| 3two.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.18Å | 0.26 | 0.15 | Mannitol dehydrogenase |
| 3wid.1.A | 8.96 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.25 | 0.15 | Glucose 1-dehydrogenase |
| 5bse.1.A | 19.05 | homo-10-mer | HHblits | X-ray | 1.70Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase |
| 5bsh.1.B | 19.05 | homo-10-mer | HHblits | X-ray | 2.10Å | 0.29 | 0.14 | Pyrroline-5-carboxylate reductase |
| 1np3.1.A | 15.38 | homo-12-mer | HHblits | X-ray | 2.00Å | 0.27 | 0.15 | Ketol-acid reductoisomerase |
| 5uyy.1.B | 17.46 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.14 | Prephenate dehydrogenase |
| 5uyy.1.A | 17.46 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.14 | Prephenate dehydrogenase |
| 5uyy.2.A | 17.46 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.14 | Prephenate dehydrogenase |
| 3wnq.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.95Å | 0.26 | 0.15 | (R)-specific carbonyl reductase |
| 3wle.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.16Å | 0.26 | 0.15 | (R)-specific carbonyl reductase |
| 3wlf.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.15 | (R)-specific carbonyl reductase |
| 3wlf.1.C | 13.64 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.15 | (R)-specific carbonyl reductase |
| 3wlf.1.D | 13.64 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.15 | (R)-specific carbonyl reductase |
| 4c4o.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.26 | 0.15 | CARBONYL REDUCTASE CPCR2 |
| 4c4o.1.B | 13.64 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.26 | 0.15 | CARBONYL REDUCTASE CPCR2 |
| 1vll.1.A | 9.09 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.15 | alanine dehydrogenase |
| 1omo.1.A | 9.09 | homo-dimer | HHblits | X-ray | 2.32Å | 0.26 | 0.15 | alanine dehydrogenase |
| 1omo.1.B | 9.09 | homo-dimer | HHblits | X-ray | 2.32Å | 0.26 | 0.15 | alanine dehydrogenase |
| 4xiy.1.A | 19.05 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.14 | Ketol-acid reductoisomerase |
| 4xiy.1.B | 19.05 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.14 | Ketol-acid reductoisomerase |
| 1hdo.1.A | 11.94 | monomer | HHblits | X-ray | 1.15Å | 0.24 | 0.15 | BILIVERDIN IX BETA REDUCTASE |
| 3wj7.1.A | 16.92 | homo-trimer | HHblits | X-ray | 2.60Å | 0.27 | 0.15 | Putative oxidoreductase |
| 3wj7.1.C | 16.92 | homo-trimer | HHblits | X-ray | 2.60Å | 0.27 | 0.15 | Putative oxidoreductase |
| 3q3c.1.A | 20.63 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.14 | Probable 3-hydroxyisobutyrate dehydrogenase |
| 3e23.1.A | 10.61 | monomer | HHblits | X-ray | 1.60Å | 0.25 | 0.15 | uncharacterized protein RPA2492 |
| 1up4.1.A | 16.42 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.24 | 0.15 | 6-PHOSPHO-BETA-GLUCOSIDASE |
| 2x4g.1.A | 11.94 | homo-dimer | HHblits | X-ray | 2.65Å | 0.24 | 0.15 | NUCLEOSIDE-DIPHOSPHATE-SUGAR EPIMERASE |
| 5nc8.1.A | 13.85 | homo-dimer | HHblits | X-ray | 3.09Å | 0.26 | 0.15 | Potassium efflux system protein |
| 5nc8.1.B | 13.85 | homo-dimer | HHblits | X-ray | 3.09Å | 0.26 | 0.15 | Potassium efflux system protein |
| 2c54.1.A | 14.93 | homo-dimer | HHblits | X-ray | 1.50Å | 0.24 | 0.15 | GDP-MANNOSE-3', 5'-EPIMERASE |
| 3e8x.1.A | 11.94 | homo-dimer | HHblits | X-ray | 2.10Å | 0.24 | 0.15 | Putative NAD-dependent epimerase/dehydratase |
| 1yve.1.A | 8.96 | homo-dimer | HHblits | X-ray | 1.65Å | 0.24 | 0.15 | ACETOHYDROXY ACID ISOMEROREDUCTASE |
| 3qsg.1.A | 14.29 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.14 | NAD-binding phosphogluconate dehydrogenase-like protein |
| 3gpi.1.A | 20.63 | monomer | HHblits | X-ray | 1.44Å | 0.28 | 0.14 | NAD-dependent epimerase/dehydratase |
| 3ktd.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.60Å | 0.27 | 0.14 | Prephenate dehydrogenase |
| 5t9e.1.A | 17.46 | homo-dimer | HHblits | X-ray | 2.03Å | 0.28 | 0.14 | Prephenate dehydrogenase 1 |
| 4wji.1.A | 12.70 | homo-dimer | HHblits | X-ray | 1.40Å | 0.28 | 0.14 | Putative cyclohexadienyl dehydrogenase and ADH prephenate dehydrogenase |
| 3hdj.1.A | 9.38 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.14 | Probable ornithine cyclodeaminase |
| 3hdj.1.B | 9.38 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.14 | Probable ornithine cyclodeaminase |
| 5t95.1.A | 17.46 | homo-dimer | HHblits | X-ray | 1.69Å | 0.28 | 0.14 | Prephenate dehydrogenase 1 |
| 3goh.1.A | 13.85 | monomer | HHblits | X-ray | 1.55Å | 0.25 | 0.15 | Alcohol dehydrogenase, zinc-containing |
| 2g5c.1.B | 19.05 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.14 | prephenate dehydrogenase |
| 2g5c.1.A | 19.05 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.14 | prephenate dehydrogenase |
| 2g5c.2.A | 19.05 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.14 | prephenate dehydrogenase |
| 3ggg.2.B | 17.19 | homo-dimer | HHblits | X-ray | 2.21Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 3ggg.1.B | 17.19 | homo-dimer | HHblits | X-ray | 2.21Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 3ggg.2.A | 17.19 | homo-dimer | HHblits | X-ray | 2.21Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 3ggg.1.A | 17.19 | homo-dimer | HHblits | X-ray | 2.21Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 3ggo.1.A | 17.19 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 3ggo.2.B | 17.19 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 3ggp.2.A | 17.19 | homo-dimer | HHblits | X-ray | 2.25Å | 0.26 | 0.14 | Prephenate dehydrogenase |
| 5n2i.1.A | 17.19 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.14 | Reduced coenzyme F420:NADP oxidoreductase |
| 4tsk.1.A | 15.63 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.26 | 0.14 | Ketol-acid reductoisomerase |
| 4gx0.1.A | 16.92 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.15 | TrkA domain protein |
| 4gx0.1.B | 16.92 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.15 | TrkA domain protein |
| 4gx0.2.A | 16.92 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.15 | TrkA domain protein |
| 4huj.1.A | 10.94 | monomer | HHblits | X-ray | 1.77Å | 0.26 | 0.14 | Uncharacterized protein |
| 2be9.1.A | 9.09 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.24 | 0.15 | Aspartate carbamoyltransferase |
| 1pg5.1.A | 9.09 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.24 | 0.15 | Aspartate carbamoyltransferase |
| 2uyy.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.25 | 0.15 | N-PAC PROTEIN |
| 3d1l.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.19Å | 0.26 | 0.14 | Putative NADP oxidoreductase BF3122 |
| 3d1l.1.B | 12.50 | homo-dimer | HHblits | X-ray | 2.19Å | 0.26 | 0.14 | Putative NADP oxidoreductase BF3122 |
| 3ko8.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.80Å | 0.23 | 0.15 | NAD-dependent epimerase/dehydratase |
| 3eyw.1.B | 12.50 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.14 | C-terminal domain of Glutathione-regulated potassium-efflux system protein kefC fused to full length Glutathione-regulated potassium-efflux system ancillary protein kefF |
| 3eyw.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.14 | C-terminal domain of Glutathione-regulated potassium-efflux system protein kefC fused to full length Glutathione-regulated potassium-efflux system ancillary protein kefF |
| 3l9x.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.14 | Glutathione-regulated potassium-efflux system protein kefC, linker, ancillary protein kefF |
| 4bv8.1.A | 10.94 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.14 | THIOMORPHOLINE-CARBOXYLATE DEHYDROGENASE |
| 4bv9.1.A | 10.94 | homo-dimer | HHblits | X-ray | 2.19Å | 0.26 | 0.14 | THIOMORPHOLINE-CARBOXYLATE DEHYDROGENASE |
| 4bv9.1.B | 10.94 | homo-dimer | HHblits | X-ray | 2.19Å | 0.26 | 0.14 | THIOMORPHOLINE-CARBOXYLATE DEHYDROGENASE |
| 3wic.1.A | 9.23 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.15 | Glucose 1-dehydrogenase |
| 3wic.1.B | 9.23 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.15 | Glucose 1-dehydrogenase |
| 1i2d.1.A | 19.67 | homo-hexamer | HHblits | X-ray | 2.81Å | 0.30 | 0.14 | ATP SULFURYLASE |
| 1m8p.1.A | 19.67 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.14 | sulfate adenylyltransferase |
| 2c20.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.70Å | 0.23 | 0.15 | UDP-GLUCOSE 4-EPIMERASE |
| 3doj.1.A | 17.74 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.14 | Dehydrogenase-like protein |
| 5aow.1.A | 9.38 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.14 | GLYOXYLATE HYDROXYPYRUVATE REDUCTASE |
| 5aow.1.B | 9.38 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.14 | GLYOXYLATE HYDROXYPYRUVATE REDUCTASE |
| 4ezb.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.14 | uncharacterized conserved protein |
| 2i99.1.A | 10.94 | homo-dimer | HHblits | X-ray | 2.60Å | 0.25 | 0.14 | Mu-crystallin homolog |
| 2i99.1.B | 10.94 | homo-dimer | HHblits | X-ray | 2.60Å | 0.25 | 0.14 | Mu-crystallin homolog |
| 4gx0.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.14 | TrkA domain protein |
| 4gx0.1.B | 12.50 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.14 | TrkA domain protein |
| 4gx0.2.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.14 | TrkA domain protein |
| 3rfx.1.A | 13.85 | homo-hexamer | HHblits | X-ray | 1.90Å | 0.24 | 0.15 | Uronate dehydrogenase |
| 3rft.1.A | 13.85 | homo-hexamer | HHblits | X-ray | 1.90Å | 0.24 | 0.15 | Uronate dehydrogenase |
| 4gx5.1.A | 17.19 | homo-tetramer | HHblits | X-ray | 3.70Å | 0.25 | 0.14 | TrkA domain protein |
| 3b1f.1.A | 17.46 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.14 | Putative prephenate dehydrogenase |
| 4g65.1.A | 12.70 | homo-dimer | HHblits | X-ray | 2.09Å | 0.26 | 0.14 | Trk system potassium uptake protein trkA |
| 4g65.1.B | 12.70 | homo-dimer | HHblits | X-ray | 2.09Å | 0.26 | 0.14 | Trk system potassium uptake protein trkA |
| 3wwy.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.25 | 0.14 | D-lactate dehydrogenase |
| 3wwy.1.B | 12.50 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.25 | 0.14 | D-lactate dehydrogenase |
| 2f1k.1.B | 15.87 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.14 | prephenate dehydrogenase |
| 2f1k.1.A | 15.87 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.14 | prephenate dehydrogenase |
| 2f1k.2.A | 15.87 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.14 | prephenate dehydrogenase |
| 1p91.1.A | 14.52 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.14 | Ribosomal RNA large subunit methyltransferase A |
| 1p91.1.B | 14.52 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.14 | Ribosomal RNA large subunit methyltransferase A |
| 5t9f.1.A | 16.13 | homo-dimer | HHblits | X-ray | 1.99Å | 0.27 | 0.14 | Prephenate dehydrogenase 1 |
| 3c24.1.A | 11.29 | homo-dimer | HHblits | X-ray | 1.62Å | 0.27 | 0.14 | Putative oxidoreductase |
| 3tri.1.A | 7.94 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.14 | Pyrroline-5-carboxylate reductase |
| 3tri.1.B | 7.94 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.14 | Pyrroline-5-carboxylate reductase |
| 5t57.1.A | 12.70 | homo-dimer | HHblits | X-ray | 1.65Å | 0.26 | 0.14 | Semialdehyde dehydrogenase NAD-binding protein |
| 3h8l.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.57Å | 0.27 | 0.14 | NADH oxidase |
| 1zej.1.A | 13.11 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.14 | 3-hydroxyacyl-CoA dehydrogenase |
| 4e21.1.A | 14.52 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.14 | 6-phosphogluconate dehydrogenase (Decarboxylating) |
| 1ko7.1.A | 23.73 | homo-trimer | HHblits | X-ray | 1.95Å | 0.31 | 0.13 | Hpr kinase/phosphatase |
| 1ko7.2.A | 23.73 | homo-trimer | HHblits | X-ray | 1.95Å | 0.31 | 0.13 | Hpr kinase/phosphatase |
| 4gvl.1.A | 17.46 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.25 | 0.14 | TrkA domain protein |
| 4gvl.1.B | 17.46 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.25 | 0.14 | TrkA domain protein |
| 4gvl.1.C | 17.46 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.25 | 0.14 | TrkA domain protein |
| 4gvl.1.D | 17.46 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.25 | 0.14 | TrkA domain protein |
| 3l4b.1.C | 12.90 | hetero-oligomer | HHblits | X-ray | 3.45Å | 0.26 | 0.14 | TrkA K+ Channel protien TM1088B |
| 2aem.1.A | 12.70 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.14 | Calcium-gated potassium channel mthK |
| 2aej.1.A | 12.70 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.14 | Calcium-gated potassium channel mthK |
| 2aej.1.B | 12.70 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.14 | Calcium-gated potassium channel mthK |
| 2ahr.1.A | 9.52 | homo-10-mer | HHblits | X-ray | 2.15Å | 0.25 | 0.14 | putative pyrroline carboxylate reductase |
| 2yjz.1.A | 14.52 | monomer | HHblits | X-ray | 2.20Å | 0.26 | 0.14 | METALLOREDUCTASE STEAP4 |
| 2vns.1.A | 17.74 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.14 | METALLOREDUCTASE STEAP3 |
| 2vq3.1.A | 17.74 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.14 | METALLOREDUCTASE STEAP3 |
| 2vq3.1.B | 17.74 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.14 | METALLOREDUCTASE STEAP3 |
| 3h2s.1.A | 8.06 | homo-dimer | HHblits | X-ray | 1.78Å | 0.26 | 0.14 | Putative NADH-flavin reductase |
| 2ag8.1.A | 16.39 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.14 | pyrroline-5-carboxylate reductase |
| 1i36.1.A | 16.95 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.13 | CONSERVED HYPOTHETICAL PROTEIN MTH1747 |
| 1i36.1.B | 16.95 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.13 | CONSERVED HYPOTHETICAL PROTEIN MTH1747 |
| 3oh8.1.A | 12.50 | monomer | HHblits | X-ray | 2.00Å | 0.22 | 0.14 | Nucleoside-diphosphate sugar epimerase (SulA family) |
| 4jgb.1.A | 11.48 | monomer | HHblits | X-ray | 2.05Å | 0.25 | 0.14 | Putative exported protein |
| 3aw9.1.A | 18.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.14 | NAD-dependent epimerase/dehydratase |
| 2i76.1.A | 8.33 | homo-dimer | HHblits | X-ray | 3.00Å | 0.23 | 0.14 | Hypothetical protein |
| 2i76.1.B | 8.33 | homo-dimer | HHblits | X-ray | 3.00Å | 0.23 | 0.14 | Hypothetical protein |
| 2rcy.1.A | 15.79 | homo-10-mer | HHblits | X-ray | 2.30Å | 0.27 | 0.13 | Pyrroline carboxylate reductase |
| 1kon.1.A | 6.78 | monomer | HHblits | X-ray | 2.20Å | 0.24 | 0.13 | Protein yebC |
| 3m2p.1.A | 6.67 | homo-dimer | HHblits | X-ray | 2.95Å | 0.22 | 0.14 | UDP-N-acetylglucosamine 4-epimerase |
| 3m2p.2.B | 6.67 | homo-dimer | HHblits | X-ray | 2.95Å | 0.22 | 0.14 | UDP-N-acetylglucosamine 4-epimerase |
| 3m2p.3.A | 6.67 | homo-dimer | HHblits | X-ray | 2.95Å | 0.22 | 0.14 | UDP-N-acetylglucosamine 4-epimerase |
| 2qrj.1.A | 18.52 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.12 | Saccharopine dehydrogenase, NAD+, L-lysine-forming |
| 2qrl.1.A | 18.52 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.12 | Saccharopine dehydrogenase, NAD+, L-lysine-forming |
| 2qrk.1.A | 18.52 | monomer | HHblits | X-ray | 1.75Å | 0.29 | 0.12 | Saccharopine dehydrogenase [NAD+, L-lysine-forming |
| 3cty.1.A | 22.00 | homo-dimer | HHblits | X-ray | 2.35Å | 0.31 | 0.11 | Thioredoxin reductase |
| 3k96.1.A | 24.49 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.11 | Glycerol-3-phosphate dehydrogenase [NAD(P)+] |
| 3k96.1.B | 24.49 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.11 | Glycerol-3-phosphate dehydrogenase [NAD(P)+] |
| 2b5w.1.A | 15.69 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.11 | glucose dehydrogenase |
| 2b5v.1.A | 15.69 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.11 | glucose dehydrogenase |
| 2vwg.1.B | 15.69 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.11 | GLUCOSE DEHYDROGENASE |
| 1onf.1.A | 24.49 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.11 | Glutathione reductase |
| 3ii4.1.A | 22.45 | homo-dimer | HHblits | X-ray | 2.42Å | 0.31 | 0.11 | Dihydrolipoyl dehydrogenase |
| 4m52.1.B | 22.45 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.11 | Dihydrolipoyl dehydrogenase |
| 1fl2.1.A | 22.00 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.11 | ALKYL HYDROPEROXIDE REDUCTASE SUBUNIT F |
| 4kms.1.A | 27.08 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.32 | 0.11 | Acetoacetyl-CoA reductase |
| 4o5q.1.A | 22.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.11 | Alkyl hydroperoxide reductase subunit F |
| 4o5u.1.A | 22.00 | homo-dimer | HHblits | X-ray | 2.65Å | 0.29 | 0.11 | Alkyl hydroperoxide reductase subunit F |
| 4ykg.1.B | 22.00 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.11 | Alkyl hydroperoxide reductase subunit F |
| 4ykf.1.B | 22.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | Alkyl hydroperoxide reductase subunit F |
| 4xvg.1.A | 22.00 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.11 | Alkyl hydroperoxide reductase subunit F |
| 1hyu.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.11 | ALKYL HYDROPEROXIDE REDUCTASE SUBUNIT F |
| 3nt6.1.A | 12.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.11 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 3nta.1.A | 12.00 | homo-dimer | HHblits | X-ray | 2.01Å | 0.28 | 0.11 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 3ia7.1.A | 16.33 | homo-dimer | HHblits | X-ray | 1.91Å | 0.30 | 0.11 | CalG4 |
| 3ia7.1.B | 16.33 | homo-dimer | HHblits | X-ray | 1.91Å | 0.30 | 0.11 | CalG4 |
| 4nfh.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.20Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 3lzw.1.A | 16.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.11 | Ferredoxin--NADP reductase 2 |
| 3lzx.1.A | 16.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.11 | Ferredoxin--NADP reductase 2 |
| 2cdc.1.A | 18.00 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.28 | 0.11 | GLUCOSE DEHYDROGENASE GLUCOSE 1-DEHYDROGENASE, DHG-1 |
| 2cd9.1.A | 18.00 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.11 | GLUCOSE DEHYDROGENASE |
| 5kj6.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.14Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 5cdt.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.70Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 5cds.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.40Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 5kje.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.26Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 5cdg.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.40Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 5kj1.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.20Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 5kcp.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.10Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 3oq6.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.20Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 1ju9.1.A | 22.92 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.11 | ALCOHOL DEHYDROGENASE |
| 1qv6.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 1axe.1.A | 22.92 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.11 | ALCOHOL DEHYDROGENASE |
| 1qlh.1.A | 22.92 | homo-dimer | HHblits | X-ray | 2.07Å | 0.31 | 0.11 | ALCOHOL DEHYDROGENASE |
| 1ee2.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.54Å | 0.31 | 0.11 | ALCOHOL DEHYDROGENASE |
| 5kjc.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.20Å | 0.31 | 0.11 | Alcohol dehydrogenase E chain |
| 1z74.1.A | 18.00 | monomer | HHblits | X-ray | 2.70Å | 0.27 | 0.11 | protein ArnA |
| 3jve.1.A | 16.67 | monomer | HHblits | X-ray | 1.34Å | 0.30 | 0.11 | DNA topoisomerase 2-binding protein 1 |
| 3i4c.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.11 | NAD-dependent alcohol dehydrogenase |
| 1jvb.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.30 | 0.11 | NAD(H)-DEPENDENT ALCOHOL DEHYDROGENASE |
| 1r37.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.11 | NAD-dependent alcohol dehydrogenase |
| 1nvg.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.11 | NAD-dependent alcohol dehydrogenase |
| 4ocg.1.A | 12.24 | homo-dimer | HHblits | X-ray | 2.75Å | 0.28 | 0.11 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 2eer.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.11 | NAD-dependent alcohol dehydrogenase |
| 1llu.1.A | 20.83 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.11 | Alcohol Dehydrogenase |
| 1a72.1.A | 23.40 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.11 | HORSE LIVER ALCOHOL DEHYDROGENASE |
| 1a71.1.A | 23.40 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.11 | LIVER ALCOHOL DEHYDROGENASE |
| 1axg.1.A | 23.40 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.11 | ALCOHOL DEHYDROGENASE |
| 3meq.1.B | 22.92 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.11 | Alcohol dehydrogenase, zinc-containing |
| 3meq.1.A | 22.92 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.11 | Alcohol dehydrogenase, zinc-containing |
| 5vj7.1.A | 14.00 | hetero-oligomer | HHblits | X-ray | 2.55Å | 0.26 | 0.11 | Oxidoreductase |
| 5jca.1.A | 14.00 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.26 | 0.11 | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) subunit alpha |
| 1n8k.1.A | 23.40 | homo-dimer | HHblits | X-ray | 1.13Å | 0.31 | 0.11 | Alcohol Dehydrogenase E chain |
| 1mgo.1.A | 23.40 | homo-dimer | HHblits | X-ray | 1.20Å | 0.31 | 0.11 | Alcohol Dehydrogenase E chain |
| 1deh.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.11 | HUMAN BETA1 ALCOHOL DEHYDROGENASE |
| 1hdz.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | ALCOHOL DEHYDROGENASE |
| 1hdy.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | ALCOHOL DEHYDROGENASE |
| 1htb.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.11 | BETA3 ALCOHOL DEHYDROGENASE |
| 3lxd.1.A | 20.83 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 4g9k.1.A | 20.83 | homo-dimer | HHblits | X-ray | 2.70Å | 0.29 | 0.11 | Rotenone-insensitive NADH-ubiquinone oxidoreductase |
| 2aaq.1.A | 18.37 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.11 | glutathione reductase |
| 4z6k.1.A | 22.92 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.11 | Alcohol dehydrogenase |
| 1mc5.1.A | 21.28 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.11 | Alcohol dehydrogenase class III chi chain |
| 3ntd.1.A | 12.24 | homo-dimer | HHblits | X-ray | 1.99Å | 0.27 | 0.11 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 2vdc.1.G | 14.00 | hetero-oligomer | HHblits | EM | 9.50Å | 0.26 | 0.11 | GLUTAMATE SYNTHASE [NADPH] SMALL CHAIN |
| 2zbw.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.11 | Thioredoxin reductase |
| 2zbw.1.B | 25.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.11 | Thioredoxin reductase |
| 3s2e.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.76Å | 0.29 | 0.11 | Zinc-containing alcohol dehydrogenase superfamily |
| 3s2e.1.C | 18.75 | homo-tetramer | HHblits | X-ray | 1.76Å | 0.29 | 0.11 | Zinc-containing alcohol dehydrogenase superfamily |
| 4g6g.1.A | 20.83 | homo-dimer | HHblits | X-ray | 2.39Å | 0.29 | 0.11 | Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial |
| 1z75.1.A | 18.37 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.11 | protein ArnA |
| 1d1s.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | ALCOHOL DEHYDROGENASE CLASS IV SIGMA CHAIN |
| 1d1t.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.29 | 0.11 | ALCOHOL DEHYDROGENASE CLASS IV SIGMA CHAIN |
| 5k1s.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.55Å | 0.29 | 0.11 | Oxidoreductase, zinc-binding dehydrogenase family |
| 5k1s.1.B | 18.75 | homo-dimer | HHblits | X-ray | 2.55Å | 0.29 | 0.11 | Oxidoreductase, zinc-binding dehydrogenase family |
| 5k1s.2.B | 18.75 | homo-dimer | HHblits | X-ray | 2.55Å | 0.29 | 0.11 | Oxidoreductase, zinc-binding dehydrogenase family |
| 5o9f.1.A | 12.24 | homo-tetramer | HHblits | X-ray | 1.64Å | 0.27 | 0.11 | Alcohol dehydrogenase |
| 5jwa.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.16Å | 0.27 | 0.11 | NADH dehydrogenase, putative |
| 1rjw.1.A | 20.83 | homo-tetramer | HHblits | X-ray | 2.35Å | 0.29 | 0.11 | Alcohol dehydrogenase |
| 1q1n.1.A | 18.75 | homo-dimer | HHblits | X-ray | 3.15Å | 0.28 | 0.11 | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region |
| 1piw.1.B | 18.75 | homo-dimer | HHblits | X-ray | 3.00Å | 0.28 | 0.11 | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region |
| 4oaq.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.86Å | 0.28 | 0.11 | R-specific carbonyl reductase |
| 4oaq.1.B | 18.75 | homo-dimer | HHblits | X-ray | 1.86Å | 0.28 | 0.11 | R-specific carbonyl reductase |
| 3grt.1.A | 16.33 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.11 | GLUTATHIONE REDUCTASE |
| 4ylf.1.B | 14.00 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.25 | 0.11 | Dihydropyrimidine dehydrogenase subunit A |
| 4ylf.2.B | 14.00 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.25 | 0.11 | Dihydropyrimidine dehydrogenase subunit A |
| 4yry.1.B | 14.00 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.25 | 0.11 | Dihydropyrimidine dehydrogenase subunit A |
| 4yry.2.B | 14.00 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.25 | 0.11 | Dihydropyrimidine dehydrogenase subunit A |
| 3p2y.1.A | 10.42 | homo-dimer | HHblits | X-ray | 1.82Å | 0.28 | 0.11 | Alanine dehydrogenase/pyridine nucleotide transhydrogenase |
| 4rqt.1.A | 14.58 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.11 | Alcohol dehydrogenase class-P |
| 4rqu.1.B | 14.58 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.11 | Alcohol Dehydrogenase |
| 5kia.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.11 | L-threonine 3-dehydrogenase |
| 4jlw.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.28 | 0.11 | Glutathione-independent formaldehyde dehydrogenase |
| 4jbi.1.A | 16.33 | homo-tetramer | HHblits | X-ray | 2.35Å | 0.27 | 0.11 | Alcohol dehydrogenase (Zinc) |
| 4jbh.1.B | 16.33 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | Alcohol dehydrogenase (Zinc) |
| 4jbg.1.A | 16.33 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.27 | 0.11 | Alcohol dehydrogenase (Zinc) |
| 1ptj.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.61Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1ptj.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.61Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1u2d.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1l7d.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.81Å | 0.28 | 0.11 | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 |
| 1l7d.1.B | 12.50 | homo-dimer | HHblits | X-ray | 1.81Å | 0.28 | 0.11 | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 |
| 1l7d.2.A | 12.50 | homo-dimer | HHblits | X-ray | 1.81Å | 0.28 | 0.11 | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 |
| 1l7d.2.B | 12.50 | homo-dimer | HHblits | X-ray | 1.81Å | 0.28 | 0.11 | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 |
| 1l7e.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.11 | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 |
| 1l7e.2.B | 12.50 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.11 | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 |
| 1xlt.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1xlt.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1xlt.2.A | 12.50 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1xlt.2.B | 12.50 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1xlt.3.A | 12.50 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1xlt.3.B | 12.50 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1u2g.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1u2d.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1u28.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 2oor.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.32Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1f8g.2.A | 12.50 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.11 | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE |
| 1f8g.2.B | 12.50 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.11 | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE |
| 2fr8.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 2fr8.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 2fsv.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 2fsv.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 2frd.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 2frd.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1nm5.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 1nm5.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 4izh.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 4j1t.2.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.37Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 4j1t.2.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.37Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 4j16.1.B | 12.50 | hetero-oligomer | HHblits | X-ray | 2.41Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 4j16.1.A | 12.50 | hetero-oligomer | HHblits | X-ray | 2.41Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 4o9u.1.E | 12.50 | hetero-oligomer | HHblits | X-ray | 6.93Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 4o9u.1.F | 12.50 | hetero-oligomer | HHblits | X-ray | 6.93Å | 0.28 | 0.11 | NAD/NADP transhydrogenase alpha subunit 1 |
| 1u3t.1.A | 19.15 | homo-dimer | HHblits | X-ray | 2.49Å | 0.30 | 0.11 | Alcohol dehydrogenase alpha chain |
| 1m6h.1.A | 19.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.11 | Glutathione-dependent formaldehyde dehydrogenase |
| 1teh.1.A | 19.15 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.11 | HUMAN CHICHI ALCOHOL DEHYDROGENASE |
| 1o8c.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.11 | YHDH |
| 1o89.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.25Å | 0.28 | 0.11 | YHDH |
| 3cos.1.A | 19.15 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.11 | Alcohol dehydrogenase 4 |
| 2fzw.1.A | 19.15 | homo-dimer | HHblits | X-ray | 1.84Å | 0.30 | 0.11 | Alcohol dehydrogenase class III chi chain |
| 3qj5.1.A | 19.15 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.11 | Alcohol dehydrogenase class-3 |
| 1uuf.1.A | 19.15 | homo-dimer | HHblits | X-ray | 1.76Å | 0.30 | 0.11 | ZINC-TYPE ALCOHOL DEHYDROGENASE-LIKE PROTEIN YAHK |
| 4dio.1.A | 10.42 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.11 | NAD(P) transhydrogenase subunit alpha part 1 |
| 4kpr.1.A | 20.41 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.11 | Thioredoxin reductase 1, cytoplasmic |
| 1u3w.1.A | 19.15 | homo-dimer | HHblits | X-ray | 1.45Å | 0.29 | 0.11 | Alcohol dehydrogenase gamma chain |
| 1e3l.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.11 | ALCOHOL DEHYDROGENASE, CLASS II |
| 1kol.1.A | 14.58 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.28 | 0.11 | formaldehyde dehydrogenase |
| 2x8g.1.A | 14.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.24 | 0.11 | THIOREDOXIN GLUTATHIONE REDUCTASE |
| 2x99.1.A | 14.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.24 | 0.11 | THIOREDOXIN GLUTATHIONE REDUCTASE |
| 5twb.1.A | 10.00 | homo-dimer | HHblits | X-ray | 1.82Å | 0.24 | 0.11 | Ferredoxin--NADP reductase |
| 5twc.1.A | 10.00 | homo-dimer | HHblits | X-ray | 2.31Å | 0.24 | 0.11 | Ferredoxin--NADP reductase |
| 2dfv.1.A | 16.67 | monomer | HHblits | X-ray | 2.05Å | 0.27 | 0.11 | Probable L-threonine 3-dehydrogenase |
| 2d8a.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.11 | Probable L-threonine 3-dehydrogenase |
| 1p0f.1.A | 19.15 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.11 | NADP-dependent ALCOHOL DEHYDROGENASE |
| 5o8h.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 1.96Å | 0.27 | 0.11 | Alcohol dehydrogenase |
| 3gfb.1.A | 14.58 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.11 | L-threonine 3-dehydrogenase |
| 3ab1.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.39Å | 0.27 | 0.11 | Ferredoxin--NADP reductase |
| 3ab1.1.B | 18.75 | homo-dimer | HHblits | X-ray | 2.39Å | 0.27 | 0.11 | Ferredoxin--NADP reductase |
| 1pl6.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.11 | Sorbitol dehydrogenase |
| 3uko.1.A | 17.02 | homo-dimer | HHblits | X-ray | 1.40Å | 0.29 | 0.11 | Alcohol dehydrogenase class-3 |
| 4jji.1.B | 17.02 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.11 | Alcohol dehydrogenase class-3 |
| 4l0q.1.A | 17.02 | homo-dimer | HHblits | X-ray | 1.95Å | 0.29 | 0.11 | Alcohol dehydrogenase class-3 |
| 3pii.1.A | 21.28 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.29 | 0.11 | Alcohol dehydrogenase |
| 1mg0.1.A | 21.74 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.10 | Alcohol Dehydrogenase E chain |
| 1adg.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.10 | ALCOHOL DEHYDROGENASE |
| 1adc.1.B | 21.74 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.10 | ALCOHOL DEHYDROGENASE |
| 7adh.1.A | 21.74 | monomer | HHblits | X-ray | 3.20Å | 0.30 | 0.10 | ISONICOTINIMIDYLATED LIVER ALCOHOL DEHYDROGENASE |
| 6adh.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.10 | HOLO-LIVER ALCOHOL DEHYDROGENASE |
| 6adh.1.B | 21.74 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.10 | HOLO-LIVER ALCOHOL DEHYDROGENASE |
| 5vj5.1.A | 21.74 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.10 | Alcohol dehydrogenase E chain |
| 5o9d.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 1.79Å | 0.27 | 0.11 | Alcohol dehydrogenase |
| 5o8q.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.22Å | 0.27 | 0.11 | Alcohol dehydrogenase |
| 2xaa.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.27 | 0.11 | SECONDARY ALCOHOL DEHYDROGENASE |
| 5vm2.1.A | 14.58 | monomer | HHblits | X-ray | 1.98Å | 0.27 | 0.11 | Alcohol dehydrogenase |
| 5vm2.2.A | 14.58 | monomer | HHblits | X-ray | 1.98Å | 0.27 | 0.11 | Alcohol dehydrogenase |
| 4dl9.1.A | 17.02 | homo-dimer | HHblits | X-ray | 1.85Å | 0.29 | 0.11 | Alcohol dehydrogenase class III |
| 1x13.1.A | 10.42 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.11 | NAD(P) transhydrogenase subunit alpha |
| 1x13.1.B | 10.42 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.11 | NAD(P) transhydrogenase subunit alpha |
| 2bru.1.B | 10.42 | hetero-oligomer | HHblits | NMR | NA | 0.27 | 0.11 | NAD(P) TRANSHYDROGENASE SUBUNIT ALPHA |
| 2bru.1.A | 10.42 | hetero-oligomer | HHblits | NMR | NA | 0.27 | 0.11 | NAD(P) TRANSHYDROGENASE SUBUNIT ALPHA |
| 1xa0.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.11 | Putative NADPH Dependent oxidoreductases |
| 1xa0.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.11 | Putative NADPH Dependent oxidoreductases |
| 4uej.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.74Å | 0.27 | 0.11 | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE |
| 4a2c.1.B | 16.67 | homo-dimer | HHblits | X-ray | 1.87Å | 0.27 | 0.11 | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE |
| 4a2c.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.87Å | 0.27 | 0.11 | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE |
| 4ueo.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.11 | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE |
| 1h2b.1.A | 17.02 | homo-tetramer | HHblits | X-ray | 1.62Å | 0.28 | 0.11 | ALCOHOL DEHYDROGENASE |
| 1pqw.1.A | 10.42 | homo-dimer | HHblits | X-ray | 2.66Å | 0.27 | 0.11 | polyketide synthase |
| 1y9a.1.A | 14.58 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.27 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 4ejm.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.27 | 0.11 | Putative zinc-binding dehydrogenase |
| 4ilk.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.11 | Starvation sensing protein rspB |
| 4ea9.1.A | 12.24 | homo-trimer | HHblits | X-ray | 0.90Å | 0.25 | 0.11 | Perosamine N-acetyltransferase |
| 1e3e.1.A | 17.02 | homo-dimer | HHblits | X-ray | 2.12Å | 0.28 | 0.11 | ALCOHOL DEHYDROGENASE, CLASS II |
| 2b83.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 1jqb.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 1.97Å | 0.26 | 0.11 | NADP-dependent Alcohol Dehydrogenase |
| 4oki.1.B | 10.42 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.11 | Phthiocerol synthesis polyketide synthase type I PpsC |
| 4oki.1.A | 10.42 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.11 | Phthiocerol synthesis polyketide synthase type I PpsC |
| 5kmp.1.A | 10.20 | homo-dimer | HHblits | X-ray | 3.20Å | 0.25 | 0.11 | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| 2qyt.1.A | 19.15 | monomer | HHblits | X-ray | 2.15Å | 0.28 | 0.11 | 2-dehydropantoate 2-reductase |
| 5vkt.1.A | 14.58 | homo-dimer | HHblits | X-ray | 1.83Å | 0.26 | 0.11 | Cinnamyl alcohol dehydrogenases (SbCAD4) |
| 5vkt.1.B | 14.58 | homo-dimer | HHblits | X-ray | 1.83Å | 0.26 | 0.11 | Cinnamyl alcohol dehydrogenases (SbCAD4) |
| 3qe3.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.26 | 0.11 | Sorbitol dehydrogenase |
| 3h4k.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.55Å | 0.25 | 0.11 | Thioredoxin glutathione reductase |
| 4cpd.1.A | 10.20 | homo-dimer | HHblits | X-ray | 2.74Å | 0.25 | 0.11 | ALCOHOL DEHYDROGENASE |
| 5tnx.1.A | 17.02 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.28 | 0.11 | Alcohol dehydrogenase zinc-binding domain protein |
| 5h81.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.11 | heteroyohimbine synthase THAS2 |
| 4b7c.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 4b7c.2.B | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 4b7c.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 4b7c.5.A | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 4b7c.6.A | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 4b7x.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 4b7x.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.11 | PROBABLE OXIDOREDUCTASE |
| 3ip1.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.26 | 0.11 | Alcohol dehydrogenase, zinc-containing |
| 5gxf.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.29Å | 0.26 | 0.11 | Acrylyl-CoA reductase AcuI |
| 4jxk.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.11 | oxidoreductase |
| 3fbg.1.A | 17.02 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.11 | putative arginate lyase |
| 1y9e.1.A | 17.02 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.27 | 0.11 | hypothetical protein yhfP |
| 1y9e.1.B | 17.02 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.27 | 0.11 | hypothetical protein yhfP |
| 1tt7.1.A | 17.02 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.11 | YHFP |
| 1tt7.1.B | 17.02 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.11 | YHFP |
| 1tt7.1.C | 17.02 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.27 | 0.11 | YHFP |
| 5gxe.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.11 | Acrylyl-CoA reductase AcuI |
| 2bll.1.A | 19.15 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.11 | PROTEIN YFBG |
| 2gah.1.A | 10.20 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.24 | 0.11 | heterotetrameric sarcosine oxidase alpha-subunit |
| 2dph.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.27Å | 0.26 | 0.11 | Formaldehyde dismutase |
| 1yqx.1.A | 14.58 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.11 | sinapyl alcohol dehydrogenase |
| 2oui.1.A | 14.89 | homo-tetramer | HHblits | X-ray | 1.77Å | 0.27 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 5fi3.1.A | 14.58 | homo-dimer | HHblits | X-ray | 1.05Å | 0.25 | 0.11 | Tetrahydroalstonine synthase |
| 5fi5.1.A | 14.58 | homo-dimer | HHblits | X-ray | 2.25Å | 0.25 | 0.11 | Tetrahydroalstonine synthase |
| 5fi5.1.B | 14.58 | homo-dimer | HHblits | X-ray | 2.25Å | 0.25 | 0.11 | Tetrahydroalstonine synthase |
| 4gkv.1.A | 10.42 | hetero-oligomer | HHblits | X-ray | 2.01Å | 0.25 | 0.11 | Alcohol dehydrogenase, propanol-preferring |
| 3fsr.1.A | 12.77 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 3fsr.1.D | 12.77 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 1qor.1.A | 17.02 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | QUINONE OXIDOREDUCTASE |
| 1zsv.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.11 | NADP-dependent leukotriene B4 12-hydroxydehydrogenase |
| 1cdo.1.A | 13.04 | homo-dimer | HHblits | X-ray | 2.05Å | 0.29 | 0.10 | ALCOHOL DEHYDROGENASE |
| 1cdo.1.B | 13.04 | homo-dimer | HHblits | X-ray | 2.05Å | 0.29 | 0.10 | ALCOHOL DEHYDROGENASE |
| 3ftn.1.A | 12.77 | homo-tetramer | HHblits | X-ray | 2.19Å | 0.27 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 4eez.1.A | 17.02 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.11 | Alcohol dehydrogenase 1 |
| 4eez.1.B | 17.02 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.11 | Alcohol dehydrogenase 1 |
| 4eex.1.A | 17.02 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | Alcohol dehydrogenase 1 |
| 4eex.1.B | 17.02 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | Alcohol dehydrogenase 1 |
| 1f8f.1.A | 17.02 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | BENZYL ALCOHOL DEHYDROGENASE |
| 1yb5.1.A | 12.77 | homo-dimer | HHblits | X-ray | 1.85Å | 0.27 | 0.11 | Quinone oxidoreductase |
| 4eaa.1.A | 12.50 | homo-trimer | HHblits | X-ray | 1.45Å | 0.25 | 0.11 | Perosamine N-acetyltransferase |
| 2h6e.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.25 | 0.11 | D-arabinose 1-dehydrogenase |
| 3fpl.1.A | 10.42 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.25 | 0.11 | NADP-dependent alcohol dehydrogenase |
| 1vj0.1.A | 14.89 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.11 | alcohol dehydrogenase, zinc-containing |
| 4dvj.1.A | 14.89 | homo-dimer | HHblits | X-ray | 1.99Å | 0.27 | 0.11 | Putative zinc-dependent alcohol dehydrogenase protein |
| 5h83.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.25Å | 0.25 | 0.11 | heteroyohimbine synthase HYS |
| 3tqh.1.A | 14.89 | monomer | HHblits | X-ray | 2.44Å | 0.26 | 0.11 | Quinone oxidoreductase |
| 4eab.1.A | 12.50 | homo-trimer | HHblits | X-ray | 1.35Å | 0.25 | 0.11 | Perosamine N-acetyltransferase |
| 2oby.1.A | 10.42 | homo-dimer | HHblits | X-ray | 3.00Å | 0.25 | 0.11 | Putative quinone oxidoreductase |
| 1h7x.1.A | 8.16 | homo-dimer | HHblits | X-ray | 2.01Å | 0.23 | 0.11 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1gt8.1.A | 8.16 | homo-dimer | HHblits | X-ray | 3.30Å | 0.23 | 0.11 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1gth.1.A | 8.16 | homo-dimer | HHblits | X-ray | 2.25Å | 0.23 | 0.11 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1h7w.2.A | 8.16 | homo-dimer | HHblits | X-ray | 1.90Å | 0.23 | 0.11 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 2x1h.1.A | 12.77 | monomer | HHblits | X-ray | 1.75Å | 0.26 | 0.11 | ZINC-BINDING ALCOHOL DEHYDROGENASE DOMAIN-CONTAINING PROTEIN 2 |
| 3wfj.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.10 | 2-dehydropantoate 2-reductase |
| 3wfi.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.10 | 2-dehydropantoate 2-reductase |
| 3wfi.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.10 | 2-dehydropantoate 2-reductase |
| 5x20.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.10 | 2-dehydropantoate 2-reductase |
| 2wek.1.A | 12.77 | monomer | HHblits | X-ray | 1.90Å | 0.26 | 0.11 | ZINC-BINDING ALCOHOL DEHYDROGENASE DOMAIN-CONTAINING PROTEIN 2 |
| 3fpc.1.A | 15.22 | homo-tetramer | HHblits | X-ray | 1.40Å | 0.27 | 0.10 | NADP-dependent alcohol dehydrogenase |
| 3nx4.1.A | 17.39 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.10 | Putative oxidoreductase |
| 3nx4.1.B | 17.39 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.10 | Putative oxidoreductase |
| 1v3t.1.A | 10.42 | homo-dimer | HHblits | X-ray | 2.30Å | 0.24 | 0.11 | leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase |
| 1v3u.1.B | 10.42 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.11 | leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase |
| 1v3u.1.A | 10.42 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.11 | leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase |
| 1e3j.1.A | 12.77 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.25 | 0.11 | NADP(H)-DEPENDENT KETOSE REDUCTASE |
| 5env.1.C | 14.89 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.25 | 0.11 | Alcohol dehydrogenase 1 |
| 5env.1.B | 14.89 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.25 | 0.11 | Alcohol dehydrogenase 1 |
| 2x7h.1.A | 12.77 | monomer | HHblits | X-ray | 1.60Å | 0.25 | 0.11 | ZINC-BINDING ALCOHOL DEHYDROGENASE DOMAIN-CONTAINING PROTEIN 2 |
| 4rvs.1.A | 10.42 | homo-dimer | HHblits | X-ray | 1.85Å | 0.24 | 0.11 | Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) |
| 2cf5.1.A | 10.64 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.11 | CINNAMYL ALCOHOL DEHYDROGENASE |
| 2cf6.1.A | 10.64 | monomer | HHblits | X-ray | 2.60Å | 0.25 | 0.11 | CINNAMYL ALCOHOL DEHYDROGENASE |
| 1bxz.1.A | 10.64 | homo-tetramer | HHblits | X-ray | 2.99Å | 0.25 | 0.11 | NADP-DEPENDENT ALCOHOL DEHYDROGENASE |
| 1kev.1.A | 13.04 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.10 | NADP-DEPENDENT ALCOHOL DEHYDROGENASE |
| 1ped.1.A | 13.04 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.27 | 0.10 | NADP-DEPENDENT ALCOHOL DEHYDROGENASE |
| 4oh1.1.A | 17.39 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.10 | L-iditol 2-dehydrogenase |
| 5lb9.1.A | 10.42 | homo-dimer | HHblits | X-ray | 2.10Å | 0.23 | 0.11 | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial |
| 5lb9.1.B | 10.42 | homo-dimer | HHblits | X-ray | 2.10Å | 0.23 | 0.11 | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial |
| 1n9g.1.A | 10.42 | hetero-oligomer | HHblits | X-ray | 1.98Å | 0.23 | 0.11 | 2,4-dienoyl-CoA reductase |
| 1n9g.1.C | 10.42 | hetero-oligomer | HHblits | X-ray | 1.98Å | 0.23 | 0.11 | 2,4-dienoyl-CoA reductase |
| 4was.2.A | 10.42 | homo-dimer | HHblits | X-ray | 1.70Å | 0.23 | 0.11 | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial |
| 1gu7.1.A | 10.42 | homo-dimer | HHblits | X-ray | 1.70Å | 0.23 | 0.11 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1,MITOCHONDRIAL |
| 1gyr.1.A | 10.42 | monomer | HHblits | X-ray | 2.60Å | 0.23 | 0.11 | 2,4-DIENOYL-COA REDUCTASE |
| 1n9g.1.B | 10.42 | hetero-oligomer | HHblits | X-ray | 1.98Å | 0.23 | 0.11 | 2,4-dienoyl-CoA reductase |
| 1n9g.1.D | 10.42 | hetero-oligomer | HHblits | X-ray | 1.98Å | 0.23 | 0.11 | 2,4-dienoyl-CoA reductase |
| 4w99.1.A | 10.42 | homo-dimer | HHblits | X-ray | 2.00Å | 0.23 | 0.11 | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial |
| 4w99.1.B | 10.42 | homo-dimer | HHblits | X-ray | 2.00Å | 0.23 | 0.11 | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial |
| 1iz0.1.A | 10.42 | monomer | HHblits | X-ray | 2.30Å | 0.23 | 0.11 | QUINONE OXIDOREDUCTASE |
| 2eih.1.A | 8.33 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.23 | 0.11 | Alcohol dehydrogenase |
| 2eih.1.B | 8.33 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.23 | 0.11 | Alcohol dehydrogenase |
| 4eye.1.A | 10.87 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.10 | Probable oxidoreductase |
| 2y05.1.A | 12.77 | monomer | HHblits | X-ray | 2.20Å | 0.25 | 0.11 | PROSTAGLANDIN REDUCTASE 1 |
| 4mkr.1.A | 14.89 | homo-dimer | HHblits | X-ray | 2.58Å | 0.25 | 0.11 | Zingiber officinale double bond reductase |
| 4nh4.1.B | 14.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.11 | Zingiber officinale double bond reductase |
| 4hfj.1.A | 17.02 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.11 | Allyl alcohol dehydrogenase |
| 5dp1.1.A | 10.87 | monomer | HHblits | X-ray | 1.85Å | 0.26 | 0.10 | CurK |
| 2j8z.1.A | 10.64 | homo-dimer | HHblits | X-ray | 2.50Å | 0.25 | 0.11 | QUINONE OXIDOREDUCTASE |
| 3hwr.1.A | 15.22 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.10 | 2-dehydropantoate 2-reductase |
| 4dup.1.A | 13.04 | homo-dimer | HHblits | X-ray | 2.45Å | 0.26 | 0.10 | quinone oxidoreductase |
| 4dup.1.B | 13.04 | homo-dimer | HHblits | X-ray | 2.45Å | 0.26 | 0.10 | quinone oxidoreductase |
| 4m98.1.A | 15.56 | homo-trimer | HHblits | X-ray | 1.67Å | 0.27 | 0.10 | Pilin glycosylation protein |
| 2nvb.1.A | 10.87 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.26 | 0.10 | NADP-dependent alcohol dehydrogenase |
| 1h0k.1.A | 10.64 | homo-dimer | HHblits | X-ray | 2.11Å | 0.24 | 0.11 | 2,4-DIENOYL-COA REDUCTASE |
| 4a0s.1.A | 8.51 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.24 | 0.11 | OCTENOYL-COA REDUCTASE/CARBOXYLASE |
| 4a10.1.B | 8.51 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.24 | 0.11 | OCTENOYL-COA REDUCTASE/CARBOXYLASE |
| 5hws.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.10 | 2-dehydropantoate 2-reductase |
| 5hws.2.B | 13.33 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.10 | 2-dehydropantoate 2-reductase |
| 2ew2.1.A | 20.93 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.10 | 2-dehydropantoate 2-reductase, putative |
| 3h2z.1.A | 13.64 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.10 | Mannitol-1-phosphate 5-dehydrogenase |
| 3uog.1.A | 8.51 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.23 | 0.11 | Alcohol dehydrogenase |
| 3m6i.1.A | 13.04 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.10 | L-arabinitol 4-dehydrogenase |
| 3jyn.1.A | 10.87 | homo-dimer | HHblits | X-ray | 2.01Å | 0.25 | 0.10 | Quinone oxidoreductase |
| 1wly.1.A | 13.04 | monomer | HHblits | X-ray | 1.30Å | 0.25 | 0.10 | 2-haloacrylate reductase |
| 4wgg.1.A | 15.22 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.10 | Double Bond Reductase |
| 3qwb.1.A | 8.70 | homo-dimer | HHblits | X-ray | 1.59Å | 0.24 | 0.10 | Probable quinone oxidoreductase |
| 3qwa.1.B | 8.70 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.10 | Probable quinone oxidoreductase |
| 3qwa.1.A | 8.70 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.10 | Probable quinone oxidoreductase |
| 3b6z.1.A | 6.38 | monomer | HHblits | X-ray | 1.88Å | 0.23 | 0.11 | Enoyl reductase |
| 3b70.1.A | 6.38 | monomer | HHblits | X-ray | 1.89Å | 0.23 | 0.11 | Enoyl reductase |
| 3ghy.1.A | 20.00 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.10 | Ketopantoate reductase protein |
| 3ghy.2.A | 20.00 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.10 | Ketopantoate reductase protein |
| 5a3j.1.A | 6.38 | monomer | HHblits | X-ray | 2.78Å | 0.22 | 0.11 | PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, CHLOROPLASTIC |
| 5a3v.1.A | 6.38 | homo-dimer | HHblits | X-ray | 2.34Å | 0.22 | 0.11 | PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, CHLOROPLASTIC |
| 3iup.1.A | 6.52 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.10 | Putative NADPH:quinone oxidoreductase |
| 3gms.1.A | 8.70 | monomer | HHblits | X-ray | 1.76Å | 0.24 | 0.10 | Putative NADPH:quinone reductase |
| 5ayv.1.A | 13.64 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.10 | 2-dehydropantoate 2-reductase |
| 5ayv.1.B | 13.64 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.10 | 2-dehydropantoate 2-reductase |
| 3do6.1.A | 29.27 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.32 | 0.09 | Formate--tetrahydrofolate ligase |
| 1txg.1.A | 13.95 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.10 | glycerol-3-phosphate dehydrogenase [NAD(P)+] |
| 3tpc.1.A | 22.50 | homo-tetramer | HHblits | X-ray | 2.34Å | 0.34 | 0.09 | Short chain alcohol dehydrogenase-related dehydrogenase |
| 4rvu.1.A | 10.87 | homo-dimer | HHblits | X-ray | 1.80Å | 0.23 | 0.10 | Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) |
| 4rvu.2.B | 10.87 | homo-dimer | HHblits | X-ray | 1.80Å | 0.23 | 0.10 | Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) |
| 3hn2.1.A | 13.95 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.10 | 2-dehydropantoate 2-reductase |
| 3pi7.1.A | 8.70 | homo-dimer | HHblits | X-ray | 1.71Å | 0.22 | 0.10 | NADH oxidoreductase |
| 4ure.1.A | 33.33 | homo-tetramer | HHblits | X-ray | 1.40Å | 0.34 | 0.09 | CYCLOHEXANOL DEHYDROGENASE |
| 3u09.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.34 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG |
| 3i83.1.A | 16.28 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.10 | 2-dehydropantoate 2-reductase |
| 3i83.1.B | 16.28 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.10 | 2-dehydropantoate 2-reductase |
| 5cej.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.34 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5cdy.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.34 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5cdy.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.34 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4u8g.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.33 | 0.09 | Putative uncharacterized protein gbs1891 |
| 4z9x.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.33 | 0.09 | Gluconate 5-dehydrogenase |
| 4nbv.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 1.64Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase putative short-chain dehydrogenases/reductases (SDR) family protein |
| 3wtc.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.33 | 0.09 | Putative oxidoreductase |
| 3wtc.1.B | 28.21 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.33 | 0.09 | Putative oxidoreductase |
| 4o5o.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 1.40Å | 0.33 | 0.09 | 3-hydroxyacyl-CoA dehydrogenase |
| 4o5o.1.B | 23.08 | homo-tetramer | HHblits | X-ray | 1.40Å | 0.33 | 0.09 | 3-hydroxyacyl-CoA dehydrogenase |
| 1q7c.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4pn3.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.33 | 0.09 | 3-hydroxyacyl-CoA dehydrogenase |
| 3tzc.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 3tzc.1.B | 25.64 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4i08.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.06Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG |
| 3tzk.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 3tzh.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4wjz.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG |
| 4wjz.1.B | 25.64 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG |
| 4wk6.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.21Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4wk6.1.B | 25.64 | homo-tetramer | HHblits | X-ray | 2.21Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 3gvc.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.33 | 0.09 | Probable short-chain type dehydrogenase/reductase |
| 2pd6.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.32 | 0.09 | Estradiol 17-beta-dehydrogenase 8 |
| 1ahi.1.D | 25.64 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE |
| 1ahh.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE |
| 5jc8.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.45Å | 0.34 | 0.09 | Putative short-chain dehydrogenase/reductase |
| 3un1.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.32 | 0.09 | Probable oxidoreductase |
| 4nbt.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.48Å | 0.34 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4nbt.1.C | 26.32 | homo-tetramer | HHblits | X-ray | 1.48Å | 0.34 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4bnt.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4afn.1.B | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4afn.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4afn.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnt.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnt.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnw.1.B | 28.21 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bny.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnw.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnw.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo2.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnz.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnz.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo1.1.B | 28.21 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo2.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo2.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo3.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo3.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo6.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo8.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo9.1.B | 28.21 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bo9.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnx.1.B | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnx.1.C | 28.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 4bnv.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.32 | 0.09 | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG |
| 3ftp.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.32 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5t5q.1.A | 27.03 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.36 | 0.08 | Short-chain dehydrogenase/reductase SDR:Glucose/ribitol dehydrogenase |
| 1k2w.1.A | 25.64 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.09 | SORBITOL DEHYDROGENASE |
| 1zk3.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.09 | R-specific alcohol dehydrogenase |
| 3ak4.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.31 | 0.09 | NADH-dependent quinuclidinone reductase |
| 3wds.1.C | 20.51 | homo-tetramer | HHblits | X-ray | 1.72Å | 0.31 | 0.09 | NADH-dependent quinuclidinone reductase |
| 4j2h.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.09 | Short chain alcohol dehydrogenase-related dehydrogenase |
| 2vou.1.A | 25.64 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.09 | 2,6-DIHYDROXYPYRIDINE HYDROXYLASE |
| 3qiv.1.A | 31.58 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.33 | 0.09 | short-chain dehydrogenase or 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4nbw.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.33 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 4nbw.1.B | 23.68 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.33 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 5end.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG |
| 3op4.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.33 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 3tzq.1.A | 28.95 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.33 | 0.09 | Short-chain type dehydrogenase/reductase |
| 3rwb.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.31 | 0.09 | Pyridoxal 4-dehydrogenase |
| 5jo9.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.31 | 0.09 | Ribitol 2-dehydrogenase |
| 3n74.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.09 | 3-ketoacyl-(acyl-carrier-protein) reductase |
| 5ha5.1.A | 28.21 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.31 | 0.09 | Brucella ovis oxidoreductase |
| 5ha5.1.D | 28.21 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.31 | 0.09 | Brucella ovis oxidoreductase |
| 4rzi.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.33 | 0.09 | 3-ketoacyl-acyl carrier protein reductase |
| 4dml.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4dmm.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.38Å | 0.31 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4dmm.1.B | 20.51 | homo-dimer | HHblits | X-ray | 2.38Å | 0.31 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 3cxr.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.33 | 0.09 | Dehydrogenase with different specificities |
| 1zem.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.09 | xylitol dehydrogenase |
| 3osu.1.A | 35.14 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.35 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 3t4x.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 3t4x.1.B | 23.08 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 3gaf.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.09 | 7-ALPHA-HYDROXYSTEROID DEHYDROGENASE |
| 3g49.1.A | 28.21 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.09 | 11-beta-hydroxysteroid dehydrogenase 1 |
| 4yae.1.A | 23.08 | homo-dimer | HHblits | X-ray | 1.60Å | 0.30 | 0.09 | C alpha-dehydrogenase |
| 5itv.1.A | 23.08 | homo-dimer | HHblits | X-ray | 2.26Å | 0.30 | 0.09 | Dihydroanticapsin 7-dehydrogenase |
| 4all.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 3gnt.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3gnt.1.B | 23.68 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2fwm.1.A | 35.14 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.34 | 0.08 | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase |
| 3m1a.1.A | 23.08 | homo-10-mer | HHblits | X-ray | 2.00Å | 0.30 | 0.09 | Putative dehydrogenase |
| 4dqx.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.09 | Probable oxidoreductase protein |
| 4alj.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4alm.1.B | 23.68 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4alm.1.C | 23.68 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4alm.1.D | 23.68 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4alm.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4aln.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4aln.1.C | 23.68 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] |
| 4cv1.1.D | 23.68 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.32 | 0.09 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 1so8.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.28 | 0.09 | 3-hydroxyacyl-CoA dehydrogenase type II |
| 2o23.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.20Å | 0.28 | 0.09 | HADH2 protein |
| 4g81.1.A | 25.64 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.30 | 0.09 | Putative hexonate dehydrogenase |
| 5l4s.1.A | 20.51 | monomer | HHblits | X-ray | 1.41Å | 0.30 | 0.09 | (-)-isopiperitenone reductase |
| 4ni5.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.30 | 0.09 | Oxidoreductase, short-chain dehydrogenase/reductase family protein |
| 4nbr.1.A | 17.95 | monomer | HHblits | X-ray | 1.35Å | 0.30 | 0.09 | Hypothetical 3-oxoacyl-(Acyl-carrier protein) reductase |
| 2zat.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.30 | 0.09 | Dehydrogenase/reductase SDR family member 4 |
| 3awd.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.32 | 0.09 | Putative polyol dehydrogenase |
| 1xg5.1.A | 28.95 | homo-tetramer | HHblits | X-ray | 1.53Å | 0.32 | 0.09 | ARPG836 |
| 1xg5.1.B | 28.95 | homo-tetramer | HHblits | X-ray | 1.53Å | 0.32 | 0.09 | ARPG836 |
| 1hxh.1.A | 29.73 | homo-tetramer | HHblits | X-ray | 1.22Å | 0.34 | 0.08 | 3BETA/17BETA-HYDROXYSTEROID DEHYDROGENASE |
| 1wnt.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.09 | L-xylulose reductase |
| 1xhl.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.30 | 0.09 | Short-chain dehydrogenase/reductase family member (5L265), putative Tropinone Reductase-II |
| 3ppi.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.09 | 3-hydroxyacyl-CoA dehydrogenase type-2 |
| 3v8b.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.30 | 0.09 | Putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier protein] reductase |
| 4trr.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.09 | Putative D-beta-hydroxybutyrate dehydrogenase |
| 2ehd.1.A | 26.32 | homo-dimer | HHblits | X-ray | 2.40Å | 0.32 | 0.09 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 2ehd.1.B | 26.32 | homo-dimer | HHblits | X-ray | 2.40Å | 0.32 | 0.09 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 4nz9.1.A | 23.68 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADPH] |
| 4fs3.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADPH] FabI |
| 3gns.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.71Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3gr6.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.28Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3pk0.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.30 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 5f1p.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.09 | PtmO8 |
| 5tii.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.09 | 3-oxoacyl-ACP reductase |
| 5tii.1.B | 23.08 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.09 | 3-oxoacyl-ACP reductase |
| 3tl3.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.30 | 0.09 | Short-chain type dehydrogenase/reductase |
| 3tl3.1.B | 23.08 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.30 | 0.09 | Short-chain type dehydrogenase/reductase |
| 1b16.1.A | 25.64 | homo-dimer | HHblits | X-ray | 1.40Å | 0.30 | 0.09 | PROTEIN (ALCOHOL DEHYDROGENASE) |
| 1b2l.1.A | 25.64 | homo-dimer | HHblits | X-ray | 1.60Å | 0.30 | 0.09 | ALCOHOL DEHYDROGENASE |
| 3rj5.1.A | 25.64 | homo-dimer | HHblits | X-ray | 1.45Å | 0.30 | 0.09 | Alcohol dehydrogenase |
| 4tkl.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.30 | 0.09 | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate |
| 3lyl.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.32 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3lyl.1.B | 23.68 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.32 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3lyl.1.D | 23.68 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.32 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3ojf.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) |
| 3ojf.1.B | 23.68 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) |
| 3ojf.1.C | 23.68 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) |
| 3ojf.1.D | 23.68 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.09 | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) |
| 4y98.1.A | 18.42 | homo-dimer | HHblits | X-ray | 2.65Å | 0.32 | 0.09 | C alpha-dehydrogenase |
| 1spx.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.09 | short-chain reductase family member (5L265) |
| 3sc4.1.A | 23.08 | homo-dimer | HHblits | X-ray | 2.50Å | 0.30 | 0.09 | Short chain dehydrogenase (A0QTM2 homolog) |
| 3ioy.1.A | 26.32 | homo-dimer | HHblits | X-ray | 1.90Å | 0.32 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 5er6.1.A | 28.95 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.32 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 4gh5.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.32 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 2hq1.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.09 | Glucose/ribitol dehydrogenase |
| 3gdg.1.B | 20.51 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.09 | Probable NADP-dependent mannitol dehydrogenase |
| 3gdg.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.09 | Probable NADP-dependent mannitol dehydrogenase |
| 3gdg.1.D | 20.51 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.09 | Probable NADP-dependent mannitol dehydrogenase |
| 3gdf.1.C | 20.51 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Probable NADP-dependent mannitol dehydrogenase |
| 3ai1.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.29 | 0.09 | NADPH-sorbose reductase |
| 4iuy.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.38Å | 0.29 | 0.09 | Short chain dehydrogenase/reductase |
| 2c73.1.A | 35.14 | homo-dimer | HHblits | X-ray | 2.20Å | 0.34 | 0.08 | AMINE OXIDASE (FLAVIN-CONTAINING) B |
| 2c75.1.A | 35.14 | homo-dimer | HHblits | X-ray | 1.70Å | 0.34 | 0.08 | AMINE OXIDASE (FLAVIN-CONTAINING) B |
| 2bk5.1.A | 35.14 | homo-dimer | HHblits | X-ray | 1.83Å | 0.34 | 0.08 | AMINE OXIDASE [FLAVIN-CONTAINING] B |
| 2xcg.1.A | 35.14 | homo-dimer | HHblits | X-ray | 1.90Å | 0.34 | 0.08 | AMINE OXIDASE [FLAVIN-CONTAINING] B |
| 1gos.1.A | 35.14 | homo-dimer | HHblits | X-ray | 3.00Å | 0.34 | 0.08 | MONOAMINE OXIDASE |
| 5jla.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.45Å | 0.31 | 0.09 | Putative short-chain dehydrogenase/reductase |
| 4yqz.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.31 | 0.09 | Putative oxidoreductase |
| 4yqz.1.D | 18.42 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.31 | 0.09 | Putative oxidoreductase |
| 1c0i.1.A | 28.95 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.09 | D-AMINO ACID OXIDASE |
| 3svt.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.09 | Short-chain type dehydrogenase/reductase |
| 3vc7.1.A | 23.08 | homo-dimer | HHblits | X-ray | 2.23Å | 0.29 | 0.09 | Putative oxidoreductase |
| 4hfr.1.A | 28.95 | homo-dimer | HHblits | X-ray | 2.73Å | 0.31 | 0.09 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4yyz.1.A | 28.95 | homo-dimer | HHblits | X-ray | 3.20Å | 0.31 | 0.09 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4yyz.1.B | 28.95 | homo-dimer | HHblits | X-ray | 3.20Å | 0.31 | 0.09 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4p38.1.A | 28.95 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.09 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 2ilt.1.A | 28.95 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.09 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 2yg3.1.A | 23.68 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.09 | PUTRESCINE OXIDASE |
| 2yg5.1.A | 23.68 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.09 | PUTRESCINE OXIDASE |
| 1u7t.1.A | 17.50 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.09 | 3-hydroxyacyl-CoA dehydrogenase type II |
| 1mg5.1.A | 22.50 | homo-dimer | HHblits | X-ray | 1.63Å | 0.27 | 0.09 | alcohol dehydrogenase |
| 3r1i.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.29 | 0.09 | Short-chain type dehydrogenase/reductase |
| 4dry.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4cql.1.A | 21.62 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.33 | 0.08 | ESTRADIOL 17-BETA-DEHYDROGENASE 8 |
| 4cql.2.D | 21.62 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.33 | 0.08 | ESTRADIOL 17-BETA-DEHYDROGENASE 8 |
| 4cql.3.D | 21.62 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.33 | 0.08 | ESTRADIOL 17-BETA-DEHYDROGENASE 8 |
| 4cql.4.D | 21.62 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.33 | 0.08 | ESTRADIOL 17-BETA-DEHYDROGENASE 8 |
| 3ihg.1.A | 26.32 | monomer | HHblits | X-ray | 2.49Å | 0.31 | 0.09 | RdmE |
| 2bgl.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.09 | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE |
| 2bgm.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.31 | 0.09 | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE |
| 2bgk.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.31 | 0.09 | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE |
| 2bgk.1.B | 26.32 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.31 | 0.09 | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE |
| 1xse.1.A | 28.95 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.09 | 11beta-hydroxysteroid dehydrogenase type 1 |
| 4nbu.1.A | 27.78 | homo-tetramer | HHblits | X-ray | 1.34Å | 0.35 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 5en4.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.52Å | 0.29 | 0.09 | 17-beta-hydroxysteroid dehydrogenase 14 |
| 1sb9.1.A | 23.08 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | wbpP |
| 3p9u.1.A | 17.95 | monomer | HHblits | X-ray | 2.81Å | 0.29 | 0.09 | TetX2 protein |
| 4w7i.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 1.98Å | 0.29 | 0.09 | 4-deoxy-L-erythro-5-hexoseulose uronate reductase A1-R' |
| 1rpn.1.A | 29.73 | monomer | HHblits | X-ray | 2.15Å | 0.33 | 0.08 | GDP-mannose 4,6-dehydratase |
| 3oml.1.A | 24.32 | homo-dimer | HHblits | X-ray | 2.15Å | 0.33 | 0.08 | Peroxisomal Multifunctional Enzyme Type 2, CG3415 |
| 4ya6.1.A | 23.68 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.09 | C alpha-dehydrogenase |
| 3nix.1.A | 18.42 | monomer | HHblits | X-ray | 2.60Å | 0.31 | 0.09 | Flavoprotein/dehydrogenase |
| 2wdz.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.29 | 0.09 | SHORT-CHAIN DEHYDROGENASE/REDUCTASE |
| 3lqf.1.D | 15.38 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.29 | 0.09 | Galactitol dehydrogenase |
| 4npc.1.A | 17.95 | homo-dimer | HHblits | X-ray | 1.75Å | 0.29 | 0.09 | Sorbitol dehydrogenase |
| 5lcx.1.A | 17.95 | monomer | HHblits | X-ray | 1.71Å | 0.29 | 0.09 | (-)-isopiperitenone reductase |
| 3e03.1.A | 23.68 | homo-dimer | HHblits | X-ray | 1.69Å | 0.31 | 0.09 | Short chain dehydrogenase |
| 4jro.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 1.92Å | 0.31 | 0.09 | FabG protein |
| 4jro.1.C | 23.68 | homo-tetramer | HHblits | X-ray | 1.92Å | 0.31 | 0.09 | FabG protein |
| 2qq5.1.A | 20.51 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.09 | Dehydrogenase/reductase SDR family member 1 |
| 4is2.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | Bile acid 3-alpha hydroxysteroid dehydrogenase |
| 4is3.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.09 | Bile acid 3-alpha hydroxysteroid dehydrogenase |
| 4is3.1.C | 20.51 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.09 | Bile acid 3-alpha hydroxysteroid dehydrogenase |
| 4q9n.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.33 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3tox.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.93Å | 0.31 | 0.09 | Short chain dehydrogenase |
| 3q6i.1.A | 27.78 | homo-dimer | HHblits | X-ray | 2.59Å | 0.35 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3q6i.1.B | 27.78 | homo-dimer | HHblits | X-ray | 2.59Å | 0.35 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3m1l.1.A | 27.78 | homo-dimer | HHblits | X-ray | 2.52Å | 0.35 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3v1t.1.A | 27.78 | homo-dimer | HHblits | X-ray | 1.88Å | 0.35 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 1zk4.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.00Å | 0.28 | 0.09 | R-specific alcohol dehydrogenase |
| 5h5x.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.09 | Putative oxidoreductase |
| 5h5x.2.B | 23.68 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.09 | Putative oxidoreductase |
| 1yde.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.09 | Retinal dehydrogenase/reductase 3 |
| 1yde.1.B | 17.95 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.09 | Retinal dehydrogenase/reductase 3 |
| 5hs6.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.02Å | 0.28 | 0.09 | 17-beta-hydroxysteroid dehydrogenase 14 |
| 4ibo.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.09 | Gluconate dehydrogenase |
| 3bxx.1.A | 23.68 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.09 | dihydroflavonol 4-reductase |
| 2nnl.1.A | 23.68 | monomer | HHblits | X-ray | 2.10Å | 0.30 | 0.09 | Dihydroflavonol 4-reductase |
| 3lz6.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 1.84Å | 0.30 | 0.09 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4dc1.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 2.82Å | 0.30 | 0.09 | Ketoacyl reductase |
| 4dc1.1.B | 21.05 | homo-tetramer | HHblits | X-ray | 2.82Å | 0.30 | 0.09 | Ketoacyl reductase |
| 5vml.1.A | 24.32 | monomer | HHblits | X-ray | 1.70Å | 0.32 | 0.08 | Acetoacetyl-CoA reductase |
| 1bdb.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.09 | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE |
| 3kvo.1.A | 17.95 | monomer | HHblits | X-ray | 2.25Å | 0.28 | 0.09 | Hydroxysteroid dehydrogenase-like protein 2 |
| 4imr.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.96Å | 0.28 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 4fn4.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.28 | 0.09 | Short chain dehydrogenase |
| 4rzh.1.A | 21.05 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4dbz.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 2.64Å | 0.30 | 0.09 | Ketoacyl reductase |
| 4dbz.1.B | 21.05 | homo-tetramer | HHblits | X-ray | 2.64Å | 0.30 | 0.09 | Ketoacyl reductase |
| 3ai3.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.30 | 0.09 | NADPH-sorbose reductase |
| 1xkq.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.32 | 0.08 | short-chain reductase family member (5D234) |
| 3rha.1.A | 27.03 | monomer | HHblits | X-ray | 2.05Å | 0.32 | 0.08 | Putrescine oxidase |
| 4lvu.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.25Å | 0.28 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 5wjs.1.A | 17.95 | homo-dimer | HHblits | X-ray | 2.15Å | 0.28 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 5feu.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.73Å | 0.28 | 0.09 | Noroxomaritidine/Norcraugsodine Reductase |
| 4h16.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.09 | Short chain alcohol dehydrogenase-related dehydrogenase |
| 4h15.1.C | 15.38 | homo-tetramer | HHblits | X-ray | 1.45Å | 0.28 | 0.09 | Short chain alcohol dehydrogenase-related dehydrogenase |
| 4h15.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 1.45Å | 0.28 | 0.09 | Short chain alcohol dehydrogenase-related dehydrogenase |
| 3k2e.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.30 | 0.09 | Enoyl-(Acyl-carrier-protein) reductase |
| 4eso.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 1.91Å | 0.30 | 0.09 | Putative oxidoreductase |
| 4eso.1.C | 23.68 | homo-tetramer | HHblits | X-ray | 1.91Å | 0.30 | 0.09 | Putative oxidoreductase |
| 3rih.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.30 | 0.09 | short chain dehydrogenase or reductase |
| 3rih.1.C | 23.68 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.30 | 0.09 | short chain dehydrogenase or reductase |
| 2pnf.1.A | 20.51 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 2bek.1.A | 20.51 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.09 | SEGREGATION PROTEIN |
| 4weo.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.30 | 0.09 | Putative acetoin(Diacetyl) reductase |
| 4opu.1.A | 23.68 | monomer | HHblits | X-ray | 2.70Å | 0.30 | 0.09 | Conserved Archaeal protein |
| 4opd.2.A | 23.68 | monomer | HHblits | X-ray | 1.81Å | 0.30 | 0.09 | Conserved Archaeal protein |
| 3atq.1.A | 23.68 | monomer | HHblits | X-ray | 1.85Å | 0.30 | 0.09 | Conserved Archaeal protein |
| 4opi.1.A | 23.68 | monomer | HHblits | X-ray | 2.24Å | 0.30 | 0.09 | Conserved Archaeal protein |
| 4opl.1.A | 23.68 | monomer | HHblits | X-ray | 2.51Å | 0.30 | 0.09 | Conserved Archaeal protein |
| 4e6p.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.32 | 0.08 | Probable sorbitol dehydrogenase (L-iditol 2-dehydrogenase) |
| 4bb6.1.A | 29.73 | homo-dimer | HHblits | X-ray | 2.55Å | 0.32 | 0.08 | CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1 |
| 4bb6.1.B | 29.73 | homo-dimer | HHblits | X-ray | 2.55Å | 0.32 | 0.08 | CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1 |
| 4c7j.1.A | 29.73 | homo-dimer | HHblits | X-ray | 2.16Å | 0.32 | 0.08 | CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1 |
| 4c7j.1.B | 29.73 | homo-dimer | HHblits | X-ray | 2.16Å | 0.32 | 0.08 | CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1 |
| 4c7k.3.A | 29.73 | homo-dimer | HHblits | X-ray | 1.91Å | 0.32 | 0.08 | CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1 |
| 3pdj.1.A | 29.73 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3pdj.1.B | 29.73 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3d5q.1.B | 29.73 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.32 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3d5q.1.A | 29.73 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.32 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3d5q.1.D | 29.73 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.32 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4r1u.1.A | 17.95 | monomer | HHblits | X-ray | 2.18Å | 0.28 | 0.09 | Cinnamoyl CoA reductase |
| 2c07.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.28 | 0.09 | 3-OXOACYL-(ACYL-CARRIER PROTEIN) REDUCTASE |
| 3i1j.1.A | 15.38 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 3i1j.1.B | 15.38 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 2z1n.1.A | 23.68 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.09 | dehydrogenase |
| 2z1n.2.A | 23.68 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.09 | dehydrogenase |
| 1iy8.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.29 | 0.09 | LEVODIONE REDUCTASE |
| 3v3n.1.A | 18.42 | monomer | HHblits | X-ray | 2.70Å | 0.29 | 0.09 | TetX2 protein |
| 3dwf.1.A | 29.73 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.08 | 11-beta-hydroxysteroid dehydrogenase 1 |
| 3dwf.1.B | 29.73 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.32 | 0.08 | 11-beta-hydroxysteroid dehydrogenase 1 |
| 4jqc.1.B | 15.79 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
| 4jqc.1.A | 15.79 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
| 3g1t.1.A | 23.68 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.09 | short chain dehydrogenase |
| 5vp5.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.34 | 0.08 | 3-oxoacyl-acyl-carrier protein reductase FabG4 |
| 3u0b.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.70Å | 0.34 | 0.08 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 3ea0.1.A | 27.78 | homo-dimer | HHblits | X-ray | 2.20Å | 0.34 | 0.08 | ATPase, ParA family |
| 3ea0.1.B | 27.78 | homo-dimer | HHblits | X-ray | 2.20Å | 0.34 | 0.08 | ATPase, ParA family |
| 1d4f.1.A | 23.68 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.29 | 0.09 | S-ADENOSYLHOMOCYSTEINE HYDROLASE |
| 3i6d.1.A | 15.79 | monomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | Protoporphyrinogen oxidase |
| 3uce.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.29 | 0.09 | Dehydrogenase |
| 3sju.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.33 | 0.08 | Keto reductase |
| 3sju.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.33 | 0.08 | Keto reductase |
| 2ptg.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Enoyl-acyl carrier reductase |
| 2yw9.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Enoyl-[acyl carrier protein] reductase |
| 2wyv.1.D | 24.32 | homo-tetramer | HHblits | X-ray | 1.86Å | 0.31 | 0.08 | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE |
| 2wyw.1.D | 24.32 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | ENOYL-[ACYL CARRIER PROTEIN] REDUCTASE |
| 3o4r.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.31 | 0.08 | Dehydrogenase/reductase SDR family member 4 |
| 2yg7.1.A | 24.32 | monomer | HHblits | X-ray | 2.75Å | 0.31 | 0.08 | PUTRESCINE OXIDASE |
| 3iah.1.A | 23.68 | homo-dimer | HHblits | X-ray | 1.83Å | 0.29 | 0.09 | Short Chain Dehydrogenase yciK |
| 1w4z.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | KETOACYL REDUCTASE |
| 1w4z.1.B | 18.42 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | KETOACYL REDUCTASE |
| 5u9p.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.29 | 0.09 | Gluconate 5-dehydrogenase |
| 5u9p.1.B | 21.05 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.29 | 0.09 | Gluconate 5-dehydrogenase |
| 3pxx.1.A | 33.33 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.33 | 0.08 | Carveol dehydrogenase |
| 4i5g.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | Alclohol dehydrogenase/short-chain dehydrogenase |
| 1ybv.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | TRIHYDROXYNAPHTHALENE REDUCTASE |
| 5tc4.1.A | 27.03 | homo-dimer | HHblits | X-ray | 1.89Å | 0.31 | 0.08 | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial |
| 5ff9.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.27 | 0.09 | Noroxomaritidine/Norcraugsodine Reductase |
| 5ff9.1.D | 15.38 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.27 | 0.09 | Noroxomaritidine/Norcraugsodine Reductase |
| 5fff.1.B | 15.38 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.27 | 0.09 | Noroxomaritidine/Norcraugsodine Reductase |
| 2ae1.1.A | 15.38 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | TROPINONE REDUCTASE-II |
| 1ipf.1.A | 15.38 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.09 | TROPINONE REDUCTASE-II |
| 3rd5.1.A | 17.95 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.09 | MYPAA.01249.C |
| 5g4k.1.B | 15.38 | hetero-oligomer | HHblits | X-ray | 1.74Å | 0.27 | 0.09 | OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN |
| 4fgs.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.76Å | 0.29 | 0.09 | Probable dehydrogenase protein |
| 3oif.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.29 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3oig.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 1.25Å | 0.29 | 0.09 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3gk3.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.33 | 0.08 | Acetoacetyl-CoA reductase |
| 5vt6.1.D | 25.00 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.33 | 0.08 | Acetoacetyl-CoA reductase |
| 3h6k.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.19Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3h6k.1.B | 27.03 | homo-dimer | HHblits | X-ray | 2.19Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3hfg.2.A | 27.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3frj.1.A | 27.03 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | Corticosteroid 11-beta-Dehydrogenase, Isozyme 1 |
| 3frj.1.B | 27.03 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | Corticosteroid 11-beta-Dehydrogenase, Isozyme 1 |
| 3d4n.1.B | 27.03 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3d3e.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 1xu7.1.A | 27.03 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase, isozyme 1 |
| 1xu7.1.C | 27.03 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase, isozyme 1 |
| 3czr.1.B | 27.03 | homo-dimer | HHblits | X-ray | 2.35Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3bzu.1.D | 27.03 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3oq1.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3oq1.1.B | 27.03 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3oq1.2.A | 27.03 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4k1l.1.D | 27.03 | homo-tetramer | HHblits | X-ray | 1.96Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4k1l.1.A | 27.03 | homo-tetramer | HHblits | X-ray | 1.96Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4k1l.1.B | 27.03 | homo-tetramer | HHblits | X-ray | 1.96Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3ch6.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.35Å | 0.31 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3all.1.A | 16.22 | monomer | HHblits | X-ray | 1.78Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 3all.2.A | 16.22 | monomer | HHblits | X-ray | 1.78Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 2b4q.1.A | 21.05 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.09 | Rhamnolipids biosynthesis 3-oxoacyl-[acyl-carrier-protein] reductase |
| 2b4q.1.B | 21.05 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.09 | Rhamnolipids biosynthesis 3-oxoacyl-[acyl-carrier-protein] reductase |
| 1zu5.1.A | 22.22 | monomer | HHblits | X-ray | 2.40Å | 0.33 | 0.08 | ftsY |
| 1zu4.1.A | 22.22 | monomer | HHblits | X-ray | 1.95Å | 0.33 | 0.08 | ftsY |
| 1d7o.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR |
| 1cwu.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | ENOYL ACP REDUCTASE |
| 3ruh.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.88Å | 0.26 | 0.09 | WbgU |
| 3ru7.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.09 | WbgU |
| 5ics.1.B | 18.42 | homo-tetramer | HHblits | X-ray | 1.52Å | 0.28 | 0.09 | 17-beta-hydroxysteroid dehydrogenase 14 |
| 5ics.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.52Å | 0.28 | 0.09 | 17-beta-hydroxysteroid dehydrogenase 14 |
| 4rgb.1.A | 35.29 | homo-dimer | HHblits | X-ray | 1.95Å | 0.37 | 0.08 | Carveol dehydrogenase |
| 1rj9.1.B | 27.78 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.33 | 0.08 | Signal recognition particle protein |
| 2cok.1.A | 16.22 | monomer | HHblits | NMR | NA | 0.30 | 0.08 | Poly [ADP-ribose] polymerase-1 |
| 4bmn.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.28 | 0.09 | ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE |
| 5u6n.2.A | 21.05 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.09 | UDP-glycosyltransferase 74F2 |
| 5u6n.1.A | 21.05 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.09 | UDP-glycosyltransferase 74F2 |
| 2a4k.1.A | 15.79 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.09 | 3-oxoacyl-[acyl carrier protein] reductase |
| 2a4k.1.B | 15.79 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.09 | 3-oxoacyl-[acyl carrier protein] reductase |
| 5epo.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.09 | 7-alpha-hydroxysteroid deydrogenase |
| 3uve.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.35 | 0.08 | Carveol dehydrogenase ((+)-trans-carveol dehydrogenase) |
| 1wvg.1.A | 29.73 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | CDP-glucose 4,6-dehydratase |
| 1wvg.1.B | 29.73 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | CDP-glucose 4,6-dehydratase |
| 3q2o.1.A | 24.32 | homo-dimer | HHblits | X-ray | 1.96Å | 0.30 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 3q2o.1.B | 24.32 | homo-dimer | HHblits | X-ray | 1.96Å | 0.30 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 4dlk.1.A | 24.32 | homo-dimer | HHblits | X-ray | 2.02Å | 0.30 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 4dlk.1.B | 24.32 | homo-dimer | HHblits | X-ray | 2.02Å | 0.30 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 3vtz.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Glucose 1-dehydrogenase |
| 3vtz.1.C | 18.92 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Glucose 1-dehydrogenase |
| 4dc0.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.81Å | 0.30 | 0.08 | Ketoacyl reductase |
| 4dc0.1.B | 21.62 | homo-tetramer | HHblits | X-ray | 2.81Å | 0.30 | 0.08 | Ketoacyl reductase |
| 2rhc.1.B | 21.62 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Actinorhodin Polyketide Ketoreductase |
| 2rhc.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Actinorhodin Polyketide Ketoreductase |
| 1enz.1.A | 21.62 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE |
| 3oey.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2irw.1.A | 30.56 | homo-dimer | HHblits | X-ray | 3.10Å | 0.32 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4bb5.1.A | 30.56 | homo-dimer | HHblits | X-ray | 2.20Å | 0.32 | 0.08 | CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1 |
| 2ffh.1.A | 27.78 | monomer | HHblits | X-ray | 3.20Å | 0.32 | 0.08 | PROTEIN (FFH) |
| 3o38.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.26 | 0.09 | Short chain dehydrogenase |
| 5vpn.1.A | 15.79 | hetero-oligomer | HHblits | X-ray | 4.22Å | 0.28 | 0.09 | Fumarate reductase flavoprotein subunit |
| 2bs2.1.A | 18.42 | hetero-oligomer | HHblits | X-ray | 1.78Å | 0.28 | 0.09 | QUINOL-FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT A |
| 1qlb.1.A | 18.42 | hetero-oligomer | HHblits | X-ray | 2.33Å | 0.28 | 0.09 | FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT |
| 3cir.1.A | 15.79 | hetero-oligomer | HHblits | X-ray | 3.65Å | 0.28 | 0.09 | Fumarate reductase flavoprotein subunit |
| 3cir.2.A | 15.79 | hetero-oligomer | HHblits | X-ray | 3.65Å | 0.28 | 0.09 | Fumarate reductase flavoprotein subunit |
| 1l0v.1.A | 15.79 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.28 | 0.09 | Fumarate reductase flavoprotein subunit |
| 1l0v.2.A | 15.79 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.28 | 0.09 | Fumarate reductase flavoprotein subunit |
| 2bs4.1.A | 18.42 | hetero-oligomer | HHblits | X-ray | 2.76Å | 0.28 | 0.09 | QUINOL-FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT A |
| 1e7p.2.D | 18.42 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.28 | 0.09 | FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT |
| 5u1g.1.A | 28.57 | hetero-oligomer | HHblits | X-ray | 3.64Å | 0.34 | 0.08 | ParA |
| 5u1g.2.B | 28.57 | hetero-oligomer | HHblits | X-ray | 3.64Å | 0.34 | 0.08 | ParA |
| 4e07.1.A | 28.57 | monomer | HHblits | X-ray | 2.90Å | 0.34 | 0.08 | Plasmid partitioning protein ParF |
| 4e09.1.A | 28.57 | homo-dimer | HHblits | X-ray | 2.99Å | 0.34 | 0.08 | Plasmid partitioning protein ParF |
| 4e03.1.A | 28.57 | monomer | HHblits | X-ray | 2.45Å | 0.34 | 0.08 | Plasmid partitioning protein ParF |
| 4e03.2.A | 28.57 | monomer | HHblits | X-ray | 2.45Å | 0.34 | 0.08 | Plasmid partitioning protein ParF |
| 4dzz.1.A | 28.57 | monomer | HHblits | X-ray | 1.80Å | 0.34 | 0.08 | Plasmid partitioning protein ParF |
| 4dzz.2.A | 28.57 | monomer | HHblits | X-ray | 1.80Å | 0.34 | 0.08 | Plasmid partitioning protein ParF |
| 4xwy.2.B | 20.51 | homo-dimer | HHblits | X-ray | 2.35Å | 0.26 | 0.09 | Sepiapterin reductase |
| 3nyw.1.A | 27.03 | homo-tetramer | HHblits | X-ray | 2.16Å | 0.30 | 0.08 | Putative oxidoreductase |
| 3nyw.1.B | 27.03 | homo-tetramer | HHblits | X-ray | 2.16Å | 0.30 | 0.08 | Putative oxidoreductase |
| 3nyw.1.C | 27.03 | homo-tetramer | HHblits | X-ray | 2.16Å | 0.30 | 0.08 | Putative oxidoreductase |
| 3nyw.1.D | 27.03 | homo-tetramer | HHblits | X-ray | 2.16Å | 0.30 | 0.08 | Putative oxidoreductase |
| 2o2s.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Enoyl-acyl carrier reductase |
| 3imf.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 1.99Å | 0.30 | 0.08 | Short chain dehydrogenase |
| 3imf.1.D | 21.62 | homo-tetramer | HHblits | X-ray | 1.99Å | 0.30 | 0.08 | Short chain dehydrogenase |
| 4nmh.1.B | 24.32 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4nmh.1.A | 24.32 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 1y5r.1.A | 24.32 | homo-dimer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | Corticosteroid 11-beta-dehydrogenase, isozyme 1 |
| 4k26.1.B | 24.32 | homo-dimer | HHblits | X-ray | 2.21Å | 0.30 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 5jy1.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.30 | 0.08 | Putative short-chain dehydrogenase/reductase |
| 3pgx.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.32 | 0.08 | carveol dehydrogenase |
| 1gz6.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.38Å | 0.32 | 0.08 | ESTRADIOL 17 BETA-DEHYDROGENASE 4 |
| 1zbq.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.19Å | 0.32 | 0.08 | 17-beta-hydroxysteroid dehydrogenase 4 |
| 3ey4.1.A | 30.56 | homo-dimer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | 11-beta-Hydroxysteroid Dehydrogenase 1 |
| 3ey4.1.B | 30.56 | homo-dimer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | 11-beta-Hydroxysteroid Dehydrogenase 1 |
| 3ey4.2.A | 30.56 | homo-dimer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | 11-beta-Hydroxysteroid Dehydrogenase 1 |
| 3ey4.2.B | 30.56 | homo-dimer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | 11-beta-Hydroxysteroid Dehydrogenase 1 |
| 1dih.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.09 | DIHYDRODIPICOLINATE REDUCTASE |
| 1drv.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.09 | DIHYDRODIPICOLINATE REDUCTASE |
| 1arz.1.C | 18.42 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.09 | DIHYDRODIPICOLINATE REDUCTASE |
| 1arz.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.09 | DIHYDRODIPICOLINATE REDUCTASE |
| 5unl.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.30 | 0.08 | 3-ketoacyl-ACP reductase |
| 2xyo.1.A | 18.92 | monomer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | TETX2 |
| 4a6n.1.A | 18.92 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | TETX2 PROTEIN |
| 4a6n.4.A | 18.92 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | TETX2 PROTEIN |
| 2xdo.1.A | 18.92 | monomer | HHblits | X-ray | 2.09Å | 0.30 | 0.08 | TETX2 PROTEIN |
| 1psd.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.32 | 0.08 | D-3-PHOSPHOGLYCERATE DEHYDROGENASE (PHOSPHOGLYCERATE DEHYDROGENASE) |
| 1psd.1.B | 25.00 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.32 | 0.08 | D-3-PHOSPHOGLYCERATE DEHYDROGENASE (PHOSPHOGLYCERATE DEHYDROGENASE) |
| 2p9g.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9g.1.B | 25.00 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9c.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.46Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9e.1.B | 25.00 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9e.1.C | 25.00 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9e.1.D | 25.00 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9e.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 2p9c.1.B | 25.00 | homo-tetramer | HHblits | X-ray | 2.46Å | 0.32 | 0.08 | D-3-phosphoglycerate dehydrogenase |
| 4x54.1.A | 15.79 | homo-dimer | HHblits | X-ray | 1.45Å | 0.28 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family |
| 1h5q.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.28 | 0.09 | NADP-DEPENDENT MANNITOL DEHYDROGENASE |
| 4o0l.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.09 | NADPH-dependent 3-quinuclidinone reductase |
| 2pla.1.A | 18.42 | homo-dimer | HHblits | X-ray | 2.51Å | 0.28 | 0.09 | Glycerol-3-phosphate dehydrogenase 1-like protein |
| 3ucx.1.A | 21.05 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.09 | Short chain dehydrogenase |
| 1nfr.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Putative oxidoreductase Rv2002 |
| 2gdz.1.A | 24.32 | homo-dimer | HHblits | X-ray | 1.65Å | 0.30 | 0.08 | NAD+-dependent 15-hydroxyprostaglandin dehydrogenase |
| 5dt9.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.66Å | 0.30 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 2o4c.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 2o4c.1.B | 27.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 2pr2.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | enoyl-ACP Reductase |
| 3fnh.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3fne.1.C | 21.62 | homo-tetramer | HHblits | X-ray | 1.98Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2x23.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 1.81Å | 0.30 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 2x22.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 2nsd.1.A | 21.62 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase |
| 1p45.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1p45.1.B | 21.62 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1p44.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1p44.2.B | 21.62 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1p44.2.C | 21.62 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2b37.2.B | 21.62 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4cod.1.C | 21.62 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 4qxm.1.B | 21.62 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4r9r.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3u49.1.A | 18.92 | homo-dimer | HHblits | X-ray | 1.75Å | 0.30 | 0.08 | Bacilysin biosynthesis oxidoreductase ywfH |
| 3u49.1.B | 18.92 | homo-dimer | HHblits | X-ray | 1.75Å | 0.30 | 0.08 | Bacilysin biosynthesis oxidoreductase ywfH |
| 3u4d.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Bacilysin biosynthesis oxidoreductase ywfH |
| 3u4c.1.B | 18.92 | homo-dimer | HHblits | X-ray | 2.03Å | 0.30 | 0.08 | Bacilysin biosynthesis oxidoreductase ywfH |
| 3nxs.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | LAO/AO transport system ATPase |
| 2ph1.1.A | 24.32 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Nucleotide-binding protein |
| 3zu2.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.10Å | 0.32 | 0.08 | PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011 |
| 4bkq.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | PUTATIVE REDUCTASE YPZ3_3519 |
| 4one.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.32 | 0.08 | 3-oxoacyl-(Acyl-carrier protein) reductase |
| 5vn2.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.08 | 3-oxoacyl-(Acyl-carrier protein) reductase |
| 5k9z.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.00Å | 0.32 | 0.08 | Putative short-chain dehydrogenase/reductase |
| 1jpj.1.A | 25.00 | monomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1jpn.1.A | 25.00 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1jpn.2.A | 25.00 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j45.1.A | 25.00 | monomer | HHblits | X-ray | 1.14Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j45.2.A | 25.00 | monomer | HHblits | X-ray | 1.14Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j46.1.A | 25.00 | monomer | HHblits | X-ray | 1.14Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j46.2.A | 25.00 | monomer | HHblits | X-ray | 1.14Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1o87.1.A | 25.00 | monomer | HHblits | X-ray | 2.10Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1o87.2.A | 25.00 | monomer | HHblits | X-ray | 2.10Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c04.1.A | 25.00 | monomer | HHblits | X-ray | 1.15Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c04.2.A | 25.00 | monomer | HHblits | X-ray | 1.15Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j7p.1.A | 25.00 | hetero-oligomer | HHblits | X-ray | 1.97Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 3ng1.1.A | 25.00 | monomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | SIGNAL SEQUENCE RECOGNITION PROTEIN FFH |
| 2cnw.1.A | 25.00 | hetero-oligomer | HHblits | X-ray | 2.39Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2cnw.3.A | 25.00 | hetero-oligomer | HHblits | X-ray | 2.39Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2xkv.1.A | 25.00 | hetero-oligomer | HHblits | EM | 13.50Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 3itd.1.A | 18.42 | homo-dimer | HHblits | X-ray | 1.89Å | 0.28 | 0.09 | 17beta-Hydroxysteroid dehydrogenase |
| 4glo.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 4gvx.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.28 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 2pff.1.A | 18.42 | hetero-oligomer | HHblits | X-ray | 4.00Å | 0.28 | 0.09 | Fatty acid synthase subunit alpha |
| 2a35.1.A | 17.95 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.09 | hypothetical protein PA4017 |
| 2a35.1.B | 17.95 | homo-dimer | HHblits | X-ray | 1.50Å | 0.25 | 0.09 | hypothetical protein PA4017 |
| 4iin.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | 3-ketoacyl-acyl carrier protein reductase (FabG) |
| 3qwf.1.B | 21.62 | homo-dimer | HHblits | X-ray | 1.88Å | 0.30 | 0.08 | 17beta-hydroxysteroid dehydrogenase |
| 3qwf.1.A | 21.62 | homo-dimer | HHblits | X-ray | 1.88Å | 0.30 | 0.08 | 17beta-hydroxysteroid dehydrogenase |
| 3is3.1.A | 21.62 | homo-dimer | HHblits | X-ray | 1.48Å | 0.30 | 0.08 | 17beta-hydroxysteroid dehydrogenase |
| 4rf3.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 1.69Å | 0.30 | 0.08 | NADPH dependent R-specific alcohol dehydrogenase |
| 5iz4.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.30 | 0.08 | Putative short-chain dehydrogenase/reductase |
| 1yxm.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | peroxisomal trans 2-enoyl CoA reductase |
| 1yxm.1.D | 24.32 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | peroxisomal trans 2-enoyl CoA reductase |
| 2ntv.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Enoyl-[ACP] reductase |
| 2ntv.1.B | 18.92 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Enoyl-[ACP] reductase |
| 1db3.1.A | 27.78 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | GDP-MANNOSE 4,6-DEHYDRATASE |
| 1vma.1.A | 22.22 | monomer | HHblits | X-ray | 1.60Å | 0.32 | 0.08 | cell division protein FtsY |
| 4bms.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.27 | 0.09 | ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE |
| 2jkc.1.A | 21.05 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | TRYPTOPHAN HALOGENASE PRNA |
| 2ar8.1.A | 21.05 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.09 | tryptophan halogenase PrnA |
| 4z43.1.A | 21.05 | homo-dimer | HHblits | X-ray | 2.29Å | 0.27 | 0.09 | Flavin-dependent tryptophan halogenase PrnA |
| 1ae1.1.A | 12.82 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.09 | TROPINONE REDUCTASE-I |
| 2uvd.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.34 | 0.08 | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE |
| 4zju.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 1.20Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1g6k.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | GLUCOSE 1-DEHYDROGENASE |
| 3p2o.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.23Å | 0.31 | 0.08 | Bifunctional protein folD |
| 4pzc.1.A | 30.56 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | 3-Hydroxyacyl-CoA dehydrogenase |
| 4pzc.2.A | 30.56 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | 3-Hydroxyacyl-CoA dehydrogenase |
| 4pzc.2.B | 30.56 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | 3-Hydroxyacyl-CoA dehydrogenase |
| 4i5d.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.09 | Alclohol dehydrogenase/short-chain dehydrogenase |
| 3lf1.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.32Å | 0.27 | 0.09 | Short Chain Oxidoreductase Q9HYA2 |
| 3lf2.1.D | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | Short Chain OxidoReductase Q9HYA2 |
| 3lf2.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | Short Chain OxidoReductase Q9HYA2 |
| 3lf2.1.B | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | Short Chain OxidoReductase Q9HYA2 |
| 3lf2.1.C | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | Short Chain OxidoReductase Q9HYA2 |
| 2x3n.1.A | 21.05 | monomer | HHblits | X-ray | 1.75Å | 0.27 | 0.09 | PROBABLE FAD-DEPENDENT MONOOXYGENASE |
| 1n7h.1.A | 24.32 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.29 | 0.08 | GDP-D-mannose-4,6-dehydratase |
| 1n7g.1.D | 24.32 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | GDP-D-mannose-4,6-dehydratase |
| 1g3r.1.A | 25.71 | monomer | HHblits | X-ray | 2.70Å | 0.34 | 0.08 | CELL DIVISION INHIBITOR |
| 1doh.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | TRIHYDROXYNAPHTHALENE REDUCTASE |
| 3ezl.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.31 | 0.08 | Acetoacetyl-CoA reductase |
| 2et6.1.A | 22.22 | monomer | HHblits | X-ray | 2.22Å | 0.31 | 0.08 | (3R)-hydroxyacyl-CoA dehydrogenase |
| 4egf.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | L-xylulose reductase |
| 3u9l.1.A | 24.32 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 3oet.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.36Å | 0.29 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 3oet.1.B | 18.92 | homo-dimer | HHblits | X-ray | 2.36Å | 0.29 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 3oet.2.A | 18.92 | homo-dimer | HHblits | X-ray | 2.36Å | 0.29 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 3oet.3.A | 18.92 | homo-dimer | HHblits | X-ray | 2.36Å | 0.29 | 0.08 | Erythronate-4-phosphate dehydrogenase |
| 1k0u.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.29 | 0.08 | S-ADENOSYL-L-HOMOCYSTEINE HYDROLASE |
| 1b3r.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | PROTEIN (S-ADENOSYLHOMOCYSTEINE HYDROLASE) |
| 4z0t.1.A | 24.32 | monomer | HHblits | X-ray | 1.50Å | 0.29 | 0.08 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 1n5d.1.A | 16.22 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | CARBONYL REDUCTASE/20BETA-HYDROXYSTEROID DEHYDROGENASE |
| 5if3.1.A | 24.32 | homo-dimer | HHblits | X-ray | 1.65Å | 0.29 | 0.08 | Short-chain dehydrogenase/reductase SDR |
| 3tsc.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.33 | 0.08 | Putative oxidoreductase |
| 2v3c.1.B | 28.57 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.33 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 2v3c.2.B | 28.57 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.33 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 2ynm.1.A | 34.29 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.33 | 0.08 | LIGHT-INDEPENDENT PROTOCHLOROPHYLLIDE REDUCTASE IRON-SULFUR ATP-BINDING PROTEIN |
| 3uic.4.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1jwa.1.A | 22.22 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.31 | 0.08 | MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN |
| 2ng1.1.A | 22.22 | monomer | HHblits | X-ray | 2.02Å | 0.31 | 0.08 | SIGNAL SEQUENCE RECOGNITION PROTEIN FFH |
| 2c03.1.A | 22.22 | monomer | HHblits | X-ray | 1.24Å | 0.31 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c03.2.A | 22.22 | monomer | HHblits | X-ray | 1.24Å | 0.31 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1ls1.1.A | 22.22 | monomer | HHblits | X-ray | 1.10Å | 0.31 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 3cio.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Tyrosine-protein kinase etk |
| 3cio.1.B | 22.22 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Tyrosine-protein kinase etk |
| 3ukq.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 3.15Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukp.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukp.1.D | 15.79 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukl.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.63Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukk.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukk.1.D | 15.79 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 5hhf.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 5hhf.1.C | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 4u8k.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukf.1.D | 15.79 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3uka.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.64Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 3ukh.1.A | 15.79 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.09 | UDP-galactopyranose mutase |
| 5aun.1.B | 41.18 | hetero-oligomer | HHblits | X-ray | 1.63Å | 0.35 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 3vx3.1.A | 41.18 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 5auq.3.A | 41.18 | homo-dimer | HHblits | X-ray | 2.53Å | 0.35 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 3vx3.1.B | 41.18 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 5axa.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.29 | 0.08 | Adenosylhomocysteinase |
| 1li4.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.01Å | 0.29 | 0.08 | S-adenosylhomocysteine hydrolase |
| 4yvf.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | Adenosylhomocysteinase |
| 3k92.1.C | 31.43 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.33 | 0.08 | NAD-specific glutamate dehydrogenase |
| 3k92.1.A | 31.43 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.33 | 0.08 | NAD-specific glutamate dehydrogenase |
| 3k92.1.B | 31.43 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.33 | 0.08 | NAD-specific glutamate dehydrogenase |
| 3wye.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 1.58Å | 0.33 | 0.08 | Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming] |
| 3v4s.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.02Å | 0.31 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 3v4s.1.B | 25.00 | homo-dimer | HHblits | X-ray | 2.02Å | 0.31 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 1enp.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | ENOYL ACYL CARRIER PROTEIN REDUCTASE |
| 1euz.1.B | 22.22 | homo-hexamer | HHblits | X-ray | 2.25Å | 0.31 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 1euz.1.A | 22.22 | homo-hexamer | HHblits | X-ray | 2.25Å | 0.31 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 1xq1.1.A | 15.79 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.09 | PUTATIVE TROPINONE REDUCATSE |
| 3t7c.1.A | 26.47 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.35 | 0.08 | Carveol dehydrogenase |
| 4b4u.1.A | 21.62 | homo-dimer | HHblits | X-ray | 1.45Å | 0.29 | 0.08 | BIFUNCTIONAL PROTEIN FOLD |
| 3gmd.1.A | 21.62 | homo-dimer | HHblits | X-ray | 2.28Å | 0.29 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 3gmd.1.B | 21.62 | homo-dimer | HHblits | X-ray | 2.28Å | 0.29 | 0.08 | Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| 4xa8.1.A | 21.62 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding |
| 5tt1.1.A | 18.92 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.08 | Putative short-chain alcohol dehydrogenase |
| 4gur.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 2.51Å | 0.33 | 0.08 | Lysine-specific histone demethylase 1B |
| 4gu0.2.B | 25.71 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.33 | 0.08 | Lysine-specific histone demethylase 1B |
| 4gu0.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.33 | 0.08 | Lysine-specific histone demethylase 1B |
| 4i5f.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | Alclohol dehydrogenase/short-chain dehydrogenase |
| 3qlj.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.08 | Short chain dehydrogenase |
| 2nlo.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.64Å | 0.31 | 0.08 | Shikimate dehydrogenase |
| 3jyo.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.00Å | 0.31 | 0.08 | Quinate/shikimate dehydrogenase |
| 5l3r.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Cell division protein FtsY homolog, chloroplastic |
| 1yb1.1.A | 15.79 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.09 | 17-beta-hydroxysteroid dehydrogenase type XI |
| 1yb1.1.B | 15.79 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.09 | 17-beta-hydroxysteroid dehydrogenase type XI |
| 1hdc.1.A | 21.62 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.08 | 3-ALPHA, 20 BETA-HYDROXYSTEROID DEHYDROGENASE |
| 1w8d.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.28 | 0.08 | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR |
| 4dyv.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.08 | Short-chain dehydrogenase/reductase SDR |
| 3rkr.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.42Å | 0.28 | 0.08 | Short chain oxidoreductase |
| 2fem.1.A | 18.92 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | Cytidylate kinase |
| 2feo.1.A | 18.92 | monomer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | Cytidylate kinase |
| 1wcv.1.A | 21.62 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.08 | SEGREGATION PROTEIN |
| 2bej.1.A | 21.62 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | SEGREGATION PROTEIN |
| 3s55.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.33 | 0.08 | Putative short-chain dehydrogenase/reductase |
| 3oid.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.33 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADPH] |
| 3oic.1.B | 28.57 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.33 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADPH] |
| 3oic.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.33 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADPH] |
| 3sj7.1.A | 31.43 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.33 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3sj7.1.B | 31.43 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.33 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 4j43.1.A | 25.00 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | PylD |
| 4j49.2.A | 25.00 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | PylD |
| 4j4b.1.A | 25.00 | monomer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | PylD |
| 4q39.1.A | 25.00 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | PYLD, pyrrolysine synthase |
| 4q3e.1.A | 25.00 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | PYLD, pyrrolysine synthase |
| 2y6r.1.A | 19.44 | monomer | HHblits | X-ray | 3.10Å | 0.31 | 0.08 | TETX2 PROTEIN |
| 5cpf.1.C | 22.22 | homo-tetramer | HHblits | X-ray | 3.41Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5cpf.1.D | 22.22 | homo-tetramer | HHblits | X-ray | 3.41Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4ohu.1.C | 22.22 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4oim.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5mtr.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4oxn.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.29Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5ugs.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2aqh.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.01Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3of2.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2ie0.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1zid.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.31 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE |
| 2ntj.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH |
| 5coq.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4dqu.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4bgi.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.31 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 2ieb.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2ied.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.14Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2ied.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.14Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2ied.1.C | 22.22 | homo-tetramer | HHblits | X-ray | 2.14Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2ied.1.D | 22.22 | homo-tetramer | HHblits | X-ray | 2.14Å | 0.31 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4rz3.1.A | 30.56 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | Site-determining protein |
| 4rz3.1.B | 30.56 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | Site-determining protein |
| 2woo.1.A | 22.22 | homo-dimer | HHblits | X-ray | 3.01Å | 0.31 | 0.08 | ATPASE GET3 |
| 3f1k.1.A | 13.16 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.09 | Uncharacterized oxidoreductase yciK |
| 3f1l.1.B | 13.16 | homo-dimer | HHblits | X-ray | 0.95Å | 0.26 | 0.09 | Uncharacterized oxidoreductase yciK |
| 3f1l.1.A | 13.16 | homo-dimer | HHblits | X-ray | 0.95Å | 0.26 | 0.09 | Uncharacterized oxidoreductase yciK |
| 3f5q.2.B | 13.16 | homo-dimer | HHblits | X-ray | 1.76Å | 0.26 | 0.09 | dehydrogenase |
| 3gz4.1.A | 13.16 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.09 | Hypothetical oxidoreductase yciK |
| 3e9q.1.A | 13.16 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.09 | Short-chain dehydrogenase |
| 3e9q.1.B | 13.16 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.09 | Short-chain dehydrogenase |
| 2zbw.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | Thioredoxin reductase |
| 2zbw.1.B | 18.92 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | Thioredoxin reductase |
| 1kdo.1.A | 18.92 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 1kdo.2.A | 18.92 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 1kdt.1.A | 18.92 | monomer | HHblits | X-ray | 1.95Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 1kdt.2.A | 18.92 | monomer | HHblits | X-ray | 1.95Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 1kdp.2.A | 18.92 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 1kdr.1.A | 18.92 | monomer | HHblits | X-ray | 2.25Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 1kdr.2.A | 18.92 | monomer | HHblits | X-ray | 2.25Å | 0.28 | 0.08 | CYTIDYLATE KINASE |
| 2cmk.1.A | 18.92 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE) |
| 1rkx.1.A | 27.78 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | CDP-glucose-4,6-dehydratase |
| 4nqz.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
| 4nr0.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
| 6ape.1.A | 22.22 | monomer | HHblits | X-ray | 1.45Å | 0.30 | 0.08 | Bifunctional protein FolD |
| 5l3r.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.33 | 0.08 | Cell division protein FtsY homolog, chloroplastic |
| 3b9q.1.A | 25.71 | monomer | HHblits | X-ray | 1.75Å | 0.33 | 0.08 | Chloroplast SRP receptor homolog, alpha subunit CPFTSY |
| 5l3q.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.33 | 0.08 | Signal recognition particle 54 kDa protein |
| 2j37.1.G | 25.71 | hetero-oligomer | HHblits | EM | 8.00Å | 0.33 | 0.08 | SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN (SRP54) |
| 2zk7.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.71Å | 0.30 | 0.08 | Glucose 1-dehydrogenase related protein |
| 2zk7.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.71Å | 0.30 | 0.08 | Glucose 1-dehydrogenase related protein |
| 1sez.1.A | 31.43 | homo-dimer | HHblits | X-ray | 2.90Å | 0.33 | 0.08 | Protoporphyrinogen oxidase, mitochondrial |
| 1sez.1.B | 31.43 | homo-dimer | HHblits | X-ray | 2.90Å | 0.33 | 0.08 | Protoporphyrinogen oxidase, mitochondrial |
| 5l3q.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.33 | 0.08 | Signal recognition particle receptor subunit alpha |
| 4ech.1.A | 19.44 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | Polyamine oxidase FMS1 |
| 2ph1.1.A | 27.78 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Nucleotide-binding protein |
| 3lu1.1.A | 21.05 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.09 | WbgU |
| 3gy0.1.A | 13.16 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.09 | dehydrogenase |
| 3f5s.1.A | 13.16 | homo-dimer | HHblits | X-ray | 1.36Å | 0.26 | 0.09 | dehydrogenase |
| 3f5s.1.B | 13.16 | homo-dimer | HHblits | X-ray | 1.36Å | 0.26 | 0.09 | dehydrogenase |
| 5va8.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.26 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 2uv8.1.A | 15.79 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.26 | 0.09 | FATTY ACID SYNTHASE SUBUNIT ALPHA (FAS2) |
| 1vl8.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.07Å | 0.28 | 0.08 | Gluconate 5-dehydrogenase |
| 1y56.1.B | 18.42 | hetero-oligomer | HHblits | X-ray | 2.86Å | 0.26 | 0.09 | sarcosine oxidase |
| 2qio.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.44Å | 0.30 | 0.08 | Enoyl-(Acyl-carrier-protein) reductase |
| 2qio.1.C | 19.44 | homo-tetramer | HHblits | X-ray | 2.44Å | 0.30 | 0.08 | Enoyl-(Acyl-carrier-protein) reductase |
| 3oje.1.A | 19.44 | homo-dimer | HHblits | X-ray | 3.02Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH) |
| 5ej2.1.A | 25.71 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.32 | 0.08 | Carveol dehydrogenase |
| 2cfc.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.32 | 0.08 | 2-(R)-HYDROXYPROPYL-COM DEHYDROGENASE |
| 1gtm.1.A | 25.71 | homo-hexamer | HHblits | X-ray | 2.20Å | 0.32 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 1gco.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | GLUCOSE DEHYDROGENASE |
| 3r3s.1.A | 13.51 | homo-tetramer | HHblits | X-ray | 1.25Å | 0.28 | 0.08 | oxidoreductase |
| 4nim.1.A | 16.22 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.08 | Versicolorin reductase |
| 3kb1.1.A | 21.62 | homo-dimer | HHblits | X-ray | 2.90Å | 0.28 | 0.08 | Nucleotide-binding protein |
| 3tdk.1.A | 16.22 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3tf5.1.A | 16.22 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 4edf.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.08Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 2dzd.1.A | 10.53 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.09 | pyruvate carboxylase |
| 3sx2.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.30 | 0.08 | Putative 3-ketoacyl-(acyl-carrier-protein) reductase |
| 4nk4.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.70Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4nk5.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2xit.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | MIPZ |
| 4dti.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4bge.1.D | 22.22 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 1bvr.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE) |
| 1bvr.1.C | 22.22 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE) |
| 2idz.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3kb1.1.A | 27.78 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Nucleotide-binding protein |
| 2woo.1.A | 31.43 | homo-dimer | HHblits | X-ray | 3.01Å | 0.32 | 0.08 | ATPASE GET3 |
| 5idx.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.28 | 0.08 | Short-chain dehydrogenase/reductase SDR |
| 4rf5.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.28 | 0.08 | NADPH dependent R-specific alcohol dehydrogenase |
| 4rf2.1.A | 18.92 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.28 | 0.08 | NADPH dependent R-specific alcohol dehydrogenase |
| 4ynt.1.A | 24.32 | monomer | HHblits | X-ray | 1.78Å | 0.28 | 0.08 | Glucose oxidase, putative |
| 2jjy.1.B | 13.89 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE |
| 2jjy.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE |
| 5l51.1.A | 16.67 | monomer | HHblits | X-ray | 2.66Å | 0.30 | 0.08 | (-)-menthone:(+)-neomenthol reductase |
| 3v2g.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 5l95.1.A | 22.22 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Ubiquitin-like modifier-activating enzyme 5 |
| 5l95.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Ubiquitin-like modifier-activating enzyme 5 |
| 1ja9.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.30 | 0.08 | 1,3,6,8-tetrahydroxynaphthalene reductase |
| 4c7o.1.B | 19.44 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE RECEPTOR FTSY |
| 2qy9.1.A | 19.44 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Cell division protein ftsY |
| 2ztu.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.32 | 0.08 | D(-)-3-hydroxybutyrate dehydrogenase |
| 1x1t.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 1.52Å | 0.32 | 0.08 | D(-)-3-hydroxybutyrate dehydrogenase |
| 2ztm.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | D(-)-3-hydroxybutyrate dehydrogenase |
| 3fwy.1.A | 31.43 | homo-dimer | HHblits | X-ray | 1.63Å | 0.32 | 0.08 | Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein |
| 3fwy.1.B | 31.43 | homo-dimer | HHblits | X-ray | 1.63Å | 0.32 | 0.08 | Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein |
| 5l3s.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.32 | 0.08 | Signal recognition particle 54 kDa protein |
| 5l3s.4.A | 25.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.32 | 0.08 | Signal recognition particle 54 kDa protein |
| 5l3v.1.A | 25.71 | monomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Signal recognition particle 54 kDa protein |
| 5l3v.2.A | 25.71 | monomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Signal recognition particle 54 kDa protein |
| 4xcv.1.A | 18.92 | homo-dimer | HHblits | X-ray | 1.40Å | 0.28 | 0.08 | NADP-dependent 2-hydroxyacid dehydrogenase |
| 5bqf.1.A | 18.92 | homo-dimer | HHblits | X-ray | 1.45Å | 0.28 | 0.08 | NADP-dependent 2-hydroxyacid dehydrogenase |
| 5tsd.1.A | 18.92 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | Probable hydroxyacid dehydrogenase protein |
| 1ion.1.A | 29.41 | monomer | HHblits | X-ray | 2.30Å | 0.34 | 0.08 | PROBABLE CELL DIVISION INHIBITOR MIND |
| 1rwb.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | Glucose 1-dehydrogenase |
| 1gee.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | GLUCOSE 1-DEHYDROGENASE |
| 3wv8.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | Hmd co-occurring protein HcgE |
| 3wv7.1.B | 25.00 | homo-trimer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | Hmd co-occurring protein HcgE |
| 3wv7.1.C | 25.00 | homo-trimer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | Hmd co-occurring protein HcgE |
| 3wv7.1.A | 25.00 | homo-trimer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | Hmd co-occurring protein HcgE |
| 3wv9.3.A | 25.00 | monomer | HHblits | X-ray | 2.75Å | 0.30 | 0.08 | Hmd co-occurring protein HcgE |
| 3wv9.4.A | 25.00 | monomer | HHblits | X-ray | 2.75Å | 0.30 | 0.08 | Hmd co-occurring protein HcgE |
| 3tfo.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.08Å | 0.30 | 0.08 | putative 3-oxoacyl-(acyl-carrier-protein) reductase |
| 3la6.1.F | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.B | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.C | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.D | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.E | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.A | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.G | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.1.H | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 3la6.2.C | 25.00 | homo-octamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Tyrosine-protein kinase wzc |
| 4ia5.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.22Å | 0.32 | 0.08 | Myosin-crossreactive antigen |
| 4ia6.1.B | 22.86 | homo-dimer | HHblits | X-ray | 1.80Å | 0.32 | 0.08 | Myosin-crossreactive antigen |
| 4nnj.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.32 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4nnj.1.C | 25.71 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.32 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5l6h.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5l6i.2.A | 25.71 | hetero-oligomer | HHblits | X-ray | 2.76Å | 0.32 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 3cmm.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.32 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 3cmm.2.A | 25.71 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.32 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 2yhs.1.A | 25.71 | monomer | HHblits | X-ray | 1.60Å | 0.32 | 0.08 | CELL DIVISION PROTEIN FTSY |
| 5j1j.1.A | 31.43 | homo-dimer | HHblits | X-ray | 1.55Å | 0.32 | 0.08 | Site-determining protein |
| 5j1j.1.B | 31.43 | homo-dimer | HHblits | X-ray | 1.55Å | 0.32 | 0.08 | Site-determining protein |
| 5jvf.1.A | 31.43 | monomer | HHblits | X-ray | 1.66Å | 0.32 | 0.08 | Site-determining protein |
| 4rlh.1.A | 10.81 | homo-tetramer | HHblits | X-ray | 2.26Å | 0.27 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3ek2.1.A | 10.81 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | Enoyl-(Acyl-carrier-protein) reductase (NADH) |
| 5j60.1.A | 24.32 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | Thioredoxin reductase |
| 1z45.1.A | 18.92 | homo-dimer | HHblits | X-ray | 1.85Å | 0.27 | 0.08 | GAL10 bifunctional protein |
| 4gcm.1.A | 16.22 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | Thioredoxin reductase |
| 1hdr.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | DIHYDROPTERIDINE REDUCTASE |
| 4a26.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | PUTATIVE C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC |
| 4a26.1.B | 18.92 | homo-dimer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | PUTATIVE C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC |
| 4m86.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2p91.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2p91.1.D | 13.89 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2xj9.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | MIPZ |
| 2xj9.1.B | 22.22 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | MIPZ |
| 4fc6.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | Peroxisomal 2,4-dienoyl-CoA reductase |
| 5ilo.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.71Å | 0.29 | 0.08 | Photoreceptor dehydrogenase, isoform C |
| 5ilg.1.B | 22.22 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | Photoreceptor dehydrogenase, isoform C |
| 5ilg.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | Photoreceptor dehydrogenase, isoform C |
| 4d0r.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.29 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 3bfv.1.A | 16.67 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.08 | Membrane protein CapA1, Protein tyrosine kinase |
| 4jmp.1.A | 16.67 | monomer | HHblits | X-ray | 1.30Å | 0.29 | 0.08 | C-terminal fragment of CapA, Protein tyrosine kinase |
| 2oze.1.A | 11.11 | homo-dimer | HHblits | X-ray | 1.83Å | 0.29 | 0.08 | Orf delta' |
| 2xxa.1.B | 19.44 | hetero-oligomer | HHblits | X-ray | 3.94Å | 0.29 | 0.08 | SRP RECEPTOR FTSY |
| 1fts.1.A | 19.44 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | FTSY |
| 4rz2.1.A | 27.78 | monomer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | Site-determining protein |
| 2ejv.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.55Å | 0.32 | 0.08 | L-threonine 3-dehydrogenase |
| 1bvu.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.32 | 0.08 | PROTEIN (GLUTAMATE DEHYDROGENASE) |
| 5uzh.1.A | 31.43 | monomer | HHblits | X-ray | 2.25Å | 0.32 | 0.08 | NafoA.00085.b |
| 2og2.1.A | 25.71 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.08 | Putative signal recognition particle receptor |
| 4c7o.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 4c7o.2.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.32 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 4i58.1.A | 25.71 | monomer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | Cyclohexylamine Oxidase |
| 4jig.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.32 | 0.08 | Dehydrogenase |
| 1zze.1.A | 18.92 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | Aldehyde reductase II |
| 1zze.2.A | 18.92 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | Aldehyde reductase II |
| 3p4s.1.A | 13.51 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.27 | 0.08 | Fumarate reductase flavoprotein subunit |
| 3p4s.2.A | 13.51 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.27 | 0.08 | Fumarate reductase flavoprotein subunit |
| 3p4q.2.A | 13.51 | hetero-oligomer | HHblits | X-ray | 3.35Å | 0.27 | 0.08 | Fumarate reductase flavoprotein subunit |
| 3p4r.2.A | 13.51 | hetero-oligomer | HHblits | X-ray | 3.05Å | 0.27 | 0.08 | Fumarate reductase flavoprotein subunit |
| 1m6v.1.A | 16.22 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.27 | 0.08 | carbamoyl phosphate synthetase large chain |
| 4u8i.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 4u8n.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 1lua.1.A | 18.92 | homo-trimer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | Methylene Tetrahydromethanopterin Dehydrogenase |
| 2o2y.1.B | 16.67 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Enoyl-acyl carrier reductase |
| 2o2y.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Enoyl-acyl carrier reductase |
| 1xwf.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | Adenosylhomocysteinase |
| 1a7a.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | S-ADENOSYLHOMOCYSTEINE HYDROLASE |
| 3nj4.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | Adenosylhomocysteinase |
| 4pfj.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | Adenosylhomocysteinase |
| 4yqy.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.38Å | 0.29 | 0.08 | Putative Dehydrogenase |
| 2oze.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.83Å | 0.29 | 0.08 | Orf delta' |
| 5l3w.1.A | 16.67 | monomer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | Signal recognition particle receptor FtsY |
| 3zms.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 2.96Å | 0.29 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A |
| 2h94.1.A | 19.44 | monomer | HHblits | X-ray | 2.90Å | 0.29 | 0.08 | Lysine-specific histone demethylase 1 |
| 2hko.1.A | 19.44 | monomer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | Lysine-specific histone demethylase 1 |
| 2xah.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.29 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1 |
| 5lhi.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 3.40Å | 0.29 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1 |
| 5l3b.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Lysine-specific histone demethylase 1A |
| 5l3d.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.29 | 0.08 | Lysine-specific histone demethylase 1A |
| 5l3c.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 3.31Å | 0.29 | 0.08 | Lysine-specific histone demethylase 1A |
| 4qej.1.A | 16.67 | homo-hexamer | HHblits | X-ray | 2.65Å | 0.29 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3prj.1.A | 16.67 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.29 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3ndb.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.31 | 0.08 | Signal recognition 54 kDa protein |
| 1j8m.1.A | 28.57 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 3uvz.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 4u8p.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 5vr8.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | UDP-glucose 6-dehydrogenase |
| 5cfz.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 1.97Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
| 1mfp.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.33Å | 0.29 | 0.08 | enoyl-[acyl-carrier-protein] reductase [Nadh] |
| 2fhs.1.A | 13.89 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | enoyl-[acyl-carrier-protein] reductase, NADH-dependent |
| 3pje.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 1dfg.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | ENOYL ACYL CARRIER PROTEIN REDUCTASE |
| 1dfi.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.29 | 0.08 | ENOYL ACYL CARRIER PROTEIN REDUCTASE |
| 1d8a.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE |
| 3pjf.1.A | 13.89 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3pjf.1.B | 13.89 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3pjd.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4cv2.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.29 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 3gn1.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | Pteridine reductase |
| 3gn2.1.D | 19.44 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.29 | 0.08 | Pteridine reductase |
| 3pvz.1.B | 13.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | UDP-N-acetylglucosamine 4,6-dehydratase |
| 3pvz.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | UDP-N-acetylglucosamine 4,6-dehydratase |
| 5u1j.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.95Å | 0.29 | 0.08 | Uncharacterized protein |
| 5u1j.1.B | 22.22 | homo-dimer | HHblits | X-ray | 2.95Å | 0.29 | 0.08 | Uncharacterized protein |
| 5u1j.2.A | 22.22 | homo-dimer | HHblits | X-ray | 2.95Å | 0.29 | 0.08 | Uncharacterized protein |
| 5u1j.2.B | 22.22 | homo-dimer | HHblits | X-ray | 2.95Å | 0.29 | 0.08 | Uncharacterized protein |
| 5k5z.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.37Å | 0.29 | 0.08 | ParA |
| 5k5z.2.A | 22.22 | homo-dimer | HHblits | X-ray | 2.37Å | 0.29 | 0.08 | ParA |
| 5k5z.2.B | 22.22 | homo-dimer | HHblits | X-ray | 2.37Å | 0.29 | 0.08 | ParA |
| 4gu1.1.A | 26.47 | monomer | HHblits | X-ray | 2.94Å | 0.34 | 0.08 | Lysine-specific histone demethylase 1B |
| 4fwe.1.A | 26.47 | monomer | HHblits | X-ray | 2.13Å | 0.34 | 0.08 | Lysine-specific histone demethylase 1B |
| 4fwf.1.A | 26.47 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.34 | 0.08 | Lysine-specific histone demethylase 1B |
| 4fwj.1.A | 26.47 | monomer | HHblits | X-ray | 2.90Å | 0.34 | 0.08 | Lysine-specific histone demethylase 1B |
| 4fwj.2.A | 26.47 | monomer | HHblits | X-ray | 2.90Å | 0.34 | 0.08 | Lysine-specific histone demethylase 1B |
| 3ndb.1.B | 29.41 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.34 | 0.08 | Signal recognition 54 kDa protein |
| 3kzv.1.A | 25.71 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | Uncharacterized oxidoreductase YIR035C |
| 1v9l.1.A | 28.57 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | glutamate dehydrogenase |
| 3alj.1.A | 17.14 | monomer | HHblits | X-ray | 1.48Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 3alm.1.A | 17.14 | monomer | HHblits | X-ray | 1.77Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 3gmc.1.A | 17.14 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 2v3c.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 2v3c.2.B | 25.71 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 2o7s.1.A | 25.71 | monomer | HHblits | X-ray | 1.78Å | 0.31 | 0.08 | Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase |
| 2yg6.1.A | 25.71 | monomer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | PUTRESCINE OXIDASE |
| 3kl4.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.31 | 0.08 | Signal recognition 54 kDa protein |
| 3orq.1.B | 16.22 | homo-dimer | HHblits | X-ray | 2.23Å | 0.27 | 0.08 | N5-Carboxyaminoimidazole ribonucleotide synthetase |
| 3orq.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.23Å | 0.27 | 0.08 | N5-Carboxyaminoimidazole ribonucleotide synthetase |
| 5b1y.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.09Å | 0.27 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 3itk.1.A | 13.51 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3itk.1.F | 13.51 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3khu.1.A | 13.51 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | UDP-glucose 6-dehydrogenase |
| 4udr.1.A | 21.05 | monomer | HHblits | X-ray | 1.60Å | 0.25 | 0.09 | GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE |
| 4udp.1.A | 21.05 | monomer | HHblits | X-ray | 1.90Å | 0.25 | 0.09 | GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE |
| 1qsg.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.29 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE |
| 1p33.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.86Å | 0.29 | 0.08 | Pteridine reductase 1 |
| 2dtd.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | Glucose 1-dehydrogenase related protein |
| 2pd3.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5l3s.1.B | 16.67 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3s.2.B | 16.67 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3s.3.B | 16.67 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3s.4.B | 16.67 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | Signal recognition particle receptor FtsY |
| 2og2.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.33 | 0.08 | Putative signal recognition particle receptor |
| 5in4.1.A | 28.57 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.31 | 0.08 | GDP-mannose 4,6 dehydratase |
| 1wxd.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | shikimate 5-dehydrogenase |
| 1wxd.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | shikimate 5-dehydrogenase |
| 2d5c.1.A | 25.71 | homo-dimer | HHblits | X-ray | 1.65Å | 0.31 | 0.08 | shikimate 5-dehydrogenase |
| 2d5c.1.B | 25.71 | homo-dimer | HHblits | X-ray | 1.65Å | 0.31 | 0.08 | shikimate 5-dehydrogenase |
| 1y8q.1.A | 28.57 | hetero-oligomer | HHblits | X-ray | 2.25Å | 0.31 | 0.08 | Ubiquitin-like 1 activating enzyme E1A |
| 1y8r.2.A | 28.57 | hetero-oligomer | HHblits | X-ray | 2.75Å | 0.31 | 0.08 | Ubiquitin-like 1 activating enzyme E1A |
| 3kyd.1.A | 28.57 | hetero-oligomer | HHblits | X-ray | 2.61Å | 0.31 | 0.08 | SUMO-activating enzyme subunit 1 |
| 3kyc.1.A | 28.57 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.31 | 0.08 | SUMO-activating enzyme subunit 1 |
| 5xtf.1.A | 13.51 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.08 | 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase |
| 4ktt.1.E | 10.81 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.27 | 0.08 | Methionine adenosyltransferase 2 subunit beta |
| 4ktt.1.F | 10.81 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.27 | 0.08 | Methionine adenosyltransferase 2 subunit beta |
| 4ktv.1.F | 10.81 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.27 | 0.08 | Methionine adenosyltransferase 2 subunit beta |
| 2ebu.1.A | 13.89 | monomer | HHblits | NMR | NA | 0.29 | 0.08 | Replication factor C subunit 1 |
| 5tjh.1.A | 16.67 | homo-hexamer | HHblits | X-ray | 2.05Å | 0.29 | 0.08 | UDP-glucose 6-dehydrogenase |
| 4j0e.1.A | 29.41 | homo-dimer | HHblits | X-ray | 1.60Å | 0.33 | 0.08 | Probable 3-hydroxyacyl-CoA dehydrogenase F54C8.1 |
| 4j0f.1.A | 29.41 | homo-dimer | HHblits | X-ray | 2.20Å | 0.33 | 0.08 | Probable 3-hydroxyacyl-CoA dehydrogenase F54C8.1 |
| 2xfo.1.A | 35.29 | homo-dimer | HHblits | X-ray | 2.10Å | 0.33 | 0.08 | AMINE OXIDASE [FLAVIN-CONTAINING] B |
| 2c76.1.A | 35.29 | homo-dimer | HHblits | X-ray | 1.70Å | 0.33 | 0.08 | AMINE OXIDASE (FLAVIN-CONTAINING) B |
| 2c72.1.A | 35.29 | homo-dimer | HHblits | X-ray | 2.00Å | 0.33 | 0.08 | AMINE OXIDASE (FLAVIN-CONTAINING) B |
| 3zq6.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.11Å | 0.31 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 3zq6.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.11Å | 0.31 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 3zq6.2.A | 20.00 | homo-dimer | HHblits | X-ray | 2.11Å | 0.31 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 3zq6.2.B | 20.00 | homo-dimer | HHblits | X-ray | 2.11Å | 0.31 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 1mxf.1.A | 25.71 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | PTERIDINE REDUCTASE 2 |
| 3zu3.1.A | 20.00 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.08 | PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011 |
| 1z6l.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Polyamine oxidase FMS1 |
| 1z6l.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Polyamine oxidase FMS1 |
| 3bi5.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Polyamine oxidase FMS1 |
| 1rsg.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | FMS1 protein |
| 1yy5.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | FMS1 protein |
| 1yy5.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | FMS1 protein |
| 1xpq.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | FMS1 protein |
| 1xpq.2.A | 20.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | FMS1 protein |
| 1xpq.2.B | 20.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | FMS1 protein |
| 4gdp.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | Polyamine oxidase FMS1 |
| 4gdp.2.B | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | Polyamine oxidase FMS1 |
| 3k8z.1.A | 25.71 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.31 | 0.08 | NAD-specific glutamate dehydrogenase |
| 3k8z.1.C | 25.71 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.31 | 0.08 | NAD-specific glutamate dehydrogenase |
| 2bek.1.A | 28.57 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.08 | SEGREGATION PROTEIN |
| 1wcv.1.A | 28.57 | homo-dimer | HHblits | X-ray | 1.60Å | 0.31 | 0.08 | SEGREGATION PROTEIN |
| 2bej.1.A | 28.57 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | SEGREGATION PROTEIN |
| 5gaf.1.7 | 20.00 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.08 | Signal recognition particle protein |
| 5gad.1.7 | 20.00 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.08 | Signal recognition particle protein Ffh |
| 5nco.1.9 | 20.00 | hetero-oligomer | HHblits | EM | 4.80Å | 0.31 | 0.08 | Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein |
| 3uov.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.05Å | 0.31 | 0.08 | OTEMO |
| 3uoz.1.A | 17.14 | monomer | HHblits | X-ray | 2.41Å | 0.31 | 0.08 | OTEMO |
| 3up4.1.A | 17.14 | monomer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | OTEMO |
| 3vdr.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.31 | 0.08 | D-3-hydroxybutyrate dehydrogenase |
| 2yz7.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.19Å | 0.31 | 0.08 | D-3-hydroxybutyrate dehydrogenase |
| 2qg4.1.B | 13.51 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | UDP-glucose 6-dehydrogenase |
| 2qg4.1.A | 13.51 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | UDP-glucose 6-dehydrogenase |
| 2hk9.1.A | 19.44 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Shikimate dehydrogenase |
| 2hk9.2.A | 19.44 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Shikimate dehydrogenase |
| 2hk9.3.A | 19.44 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Shikimate dehydrogenase |
| 2hk9.4.A | 19.44 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Shikimate dehydrogenase |
| 4fd3.1.A | 19.44 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4hbg.1.A | 19.44 | homo-dimer | HHblits | X-ray | 2.27Å | 0.29 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 4ohv.1.A | 16.67 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | Protein clpf-1 |
| 3oec.1.A | 23.53 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.33 | 0.08 | Carveol dehydrogenase (MythA.01326.c, A0R518 homolog) |
| 5k5z.1.A | 29.41 | homo-dimer | HHblits | X-ray | 2.37Å | 0.33 | 0.08 | ParA |
| 5k5z.2.A | 29.41 | homo-dimer | HHblits | X-ray | 2.37Å | 0.33 | 0.08 | ParA |
| 5k5z.2.B | 29.41 | homo-dimer | HHblits | X-ray | 2.37Å | 0.33 | 0.08 | ParA |
| 4cr6.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | N-ACYLMANNOSAMINE 1-DEHYDROGENASE |
| 4rz2.1.A | 25.71 | monomer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | Site-determining protein |
| 5nco.1.c | 25.71 | hetero-oligomer | HHblits | EM | 4.80Å | 0.31 | 0.08 | Signal recognition particle receptor FtsY |
| 2xxa.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 3.94Å | 0.31 | 0.08 | SRP RECEPTOR FTSY |
| 2qy9.1.A | 25.71 | monomer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | Cell division protein ftsY |
| 1fts.1.A | 25.71 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | FTSY |
| 1dhr.1.A | 10.81 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.08 | DIHYDROPTERIDINE REDUCTASE |
| 4zgw.1.A | 19.44 | homo-dimer | HHblits | X-ray | 1.47Å | 0.28 | 0.08 | short-chain dehydrogenase/reductase |
| 4zgw.1.B | 19.44 | homo-dimer | HHblits | X-ray | 1.47Å | 0.28 | 0.08 | short-chain dehydrogenase/reductase |
| 2ved.1.A | 13.89 | homo-octamer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | MEMBRANE PROTEIN CAPA1, PROTEIN TYROSINE KINASE |
| 4jlv.1.A | 13.89 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.08 | C-terminal fragment of Membrane protein CapA1, Putative uncharacterized protein capB1 |
| 3t4e.1.A | 11.11 | homo-dimer | HHblits | X-ray | 1.95Å | 0.28 | 0.08 | Quinate/shikimate dehydrogenase |
| 3f4b.1.A | 23.53 | homo-tetramer | HHblits | X-ray | 2.49Å | 0.33 | 0.08 | Enoyl-acyl carrier protein reductase |
| 4ue5.1.D | 26.47 | hetero-oligomer | HHblits | EM | 9.00Å | 0.33 | 0.08 | SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN |
| 3jaj.45.A | 26.47 | monomer | HHblits | EM | NA | 0.33 | 0.08 | SRP54 |
| 3jan.45.A | 26.47 | monomer | HHblits | EM | NA | 0.33 | 0.08 | SRP54 |
| 4ii3.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4ii2.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5knl.2.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5knl.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5um6.1.A | 22.86 | monomer | HHblits | X-ray | 2.79Å | 0.30 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4ak9.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | CPFTSY |
| 5l3r.1.A | 28.57 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Signal recognition particle 54 kDa protein, chloroplastic |
| 1b3b.1.A | 25.71 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.30 | 0.08 | PROTEIN (GLUTAMATE DEHYDROGENASE) |
| 2tmg.1.A | 25.71 | homo-hexamer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | PROTEIN (GLUTAMATE DEHYDROGENASE) |
| 1b26.1.A | 25.71 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 3syn.2.A | 25.71 | hetero-oligomer | HHblits | X-ray | 3.06Å | 0.30 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.1.B | 25.71 | homo-dimer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.3.B | 25.71 | homo-dimer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.4.A | 25.71 | homo-dimer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px3.1.A | 25.71 | homo-dimer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.1.A | 25.71 | homo-dimer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | Flagellar biosynthesis protein flhF |
| 1vma.1.A | 25.71 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | cell division protein FtsY |
| 4r1s.1.A | 19.44 | monomer | HHblits | X-ray | 1.60Å | 0.28 | 0.08 | cinnamoyl CoA reductase |
| 4r1t.1.A | 19.44 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | cinnamoyl CoA reductase |
| 3f8d.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.40Å | 0.28 | 0.08 | Thioredoxin reductase (TrxB-3) |
| 5fyd.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.08 | OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN |
| 5fyd.1.B | 16.67 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.08 | OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN |
| 5gt9.1.A | 16.67 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 5gt9.1.B | 16.67 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 5l3v.1.A | 25.71 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Signal recognition particle 54 kDa protein |
| 5l3v.2.A | 25.71 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Signal recognition particle 54 kDa protein |
| 1hwy.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 1hwz.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 5l3s.1.A | 25.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle 54 kDa protein |
| 5l3s.4.A | 25.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle 54 kDa protein |
| 3md0.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.45Å | 0.30 | 0.08 | Arginine/ornithine transport system ATPase |
| 2aqi.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5gad.1.7 | 20.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.08 | Signal recognition particle protein Ffh |
| 4c7o.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 4c7o.2.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 5gaf.1.7 | 20.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.08 | Signal recognition particle protein |
| 3fmi.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.18Å | 0.30 | 0.08 | Dethiobiotin synthetase |
| 3fmi.1.B | 22.86 | homo-dimer | HHblits | X-ray | 2.18Å | 0.30 | 0.08 | Dethiobiotin synthetase |
| 3fmf.2.A | 22.86 | homo-dimer | HHblits | X-ray | 2.05Å | 0.30 | 0.08 | Dethiobiotin synthetase |
| 1uay.1.A | 26.47 | homo-tetramer | HHblits | X-ray | 1.40Å | 0.33 | 0.08 | Type II 3-hydroxyacyl-CoA dehydrogenase |
| 3v2h.1.A | 23.53 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.33 | 0.08 | D-beta-hydroxybutyrate dehydrogenase |
| 4v02.1.A | 29.41 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.33 | 0.08 | SITE-DETERMINING PROTEIN |
| 5i9m.2.D | 11.11 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.28 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 4bku.1.C | 11.11 | homo-tetramer | HHblits | X-ray | 1.84Å | 0.28 | 0.08 | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] |
| 5i8w.1.C | 11.11 | homo-tetramer | HHblits | X-ray | 1.63Å | 0.28 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5koi.1.B | 19.44 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5koi.1.A | 19.44 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5koi.4.A | 19.44 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 2a9f.1.A | 18.92 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.08 | putative malic enzyme ((S)-malate:NAD+ oxidoreductase (decarboxylating)) |
| 2a9f.1.B | 18.92 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.08 | putative malic enzyme ((S)-malate:NAD+ oxidoreductase (decarboxylating)) |
| 3lxd.1.A | 22.86 | monomer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 4uir.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.75Å | 0.30 | 0.08 | OLEATE HYDRATASE |
| 4uir.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.75Å | 0.30 | 0.08 | OLEATE HYDRATASE |
| 3wtb.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | Putative oxidoreductase |
| 1zud.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 1.98Å | 0.30 | 0.08 | Adenylyltransferase thiF |
| 4mow.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.30 | 0.08 | Glucose 1-dehydrogenase |
| 4mow.1.D | 20.00 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.30 | 0.08 | Glucose 1-dehydrogenase |
| 4rz3.1.A | 25.71 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Site-determining protein |
| 4rz3.1.B | 25.71 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Site-determining protein |
| 1y0p.1.A | 25.71 | monomer | HHblits | X-ray | 1.50Å | 0.30 | 0.08 | Fumarate reductase flavoprotein subunit |
| 1q9i.1.A | 25.71 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | flavocytochrome c3 |
| 5nco.1.c | 20.00 | hetero-oligomer | HHblits | EM | 4.80Å | 0.30 | 0.08 | Signal recognition particle receptor FtsY |
| 1j8y.1.A | 23.53 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 3grk.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.35Å | 0.28 | 0.08 | Enoyl-(acyl-carrier-protein) reductase (NADH) |
| 3h4v.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | Pteridine reductase 1 |
| 3h4v.2.C | 22.22 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | Pteridine reductase 1 |
| 3h4v.2.D | 22.22 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | Pteridine reductase 1 |
| 2bfo.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | PTERIDINE REDUCTASE 1 |
| 1w0c.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | PTERIDINE REDUCTASE |
| 1w0c.1.B | 22.22 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | PTERIDINE REDUCTASE |
| 1w0c.1.C | 22.22 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | PTERIDINE REDUCTASE |
| 1e7w.1.A | 22.22 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.28 | 0.08 | PTERIDINE REDUCTASE |
| 2nm0.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 1.99Å | 0.28 | 0.08 | Probable 3-oxacyl-(Acyl-carrier-protein) reductase |
| 3wxb.1.A | 16.22 | homo-dimer | HHblits | X-ray | 1.98Å | 0.26 | 0.08 | Uncharacterized protein |
| 3wxb.1.B | 16.22 | homo-dimer | HHblits | X-ray | 1.98Å | 0.26 | 0.08 | Uncharacterized protein |
| 4rdh.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | tRNA threonylcarbamoyladenosine dehydratase |
| 4d7a.1.A | 25.71 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | TRNA THREONYLCARBAMOYLADENOSINE DEHYDRATASE |
| 4d79.1.A | 25.71 | homo-dimer | HHblits | X-ray | 1.77Å | 0.30 | 0.08 | TRNA THREONYLCARBAMOYLADENOSINE DEHYDRATASE |
| 4yed.1.A | 25.71 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | tRNA threonylcarbamoyladenosine dehydratase |
| 4c5o.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | FLAVIN MONOOXYGENASE |
| 4a5l.1.A | 28.57 | homo-dimer | HHblits | X-ray | 1.66Å | 0.30 | 0.08 | THIOREDOXIN REDUCTASE |
| 4a5l.1.B | 28.57 | homo-dimer | HHblits | X-ray | 1.66Å | 0.30 | 0.08 | THIOREDOXIN REDUCTASE |
| 4ccr.1.A | 28.57 | homo-dimer | HHblits | X-ray | 2.28Å | 0.30 | 0.08 | THIOREDOXIN REDUCTASE |
| 1xdb.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 1rw4.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 1m34.1.E | 25.71 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Nitrogenase Iron Protein 1 |
| 1m1y.1.E | 25.71 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | nitrogenase IRON protein 1 |
| 1m1y.1.F | 25.71 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | nitrogenase IRON protein 1 |
| 2afi.1.E | 25.71 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 2afh.1.E | 25.71 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 2afh.1.F | 25.71 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 1g5p.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g5p.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g1m.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g1m.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1fp6.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.15Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1nip.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1nip.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g20.1.E | 25.71 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g21.1.H | 25.71 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g21.1.E | 25.71 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 4zn0.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Thioredoxin reductase |
| 4zn0.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Thioredoxin reductase |
| 4c7o.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE RECEPTOR FTSY |
| 4wzb.1.F | 26.47 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Nitrogenase iron protein 1 |
| 4wzb.1.E | 26.47 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Nitrogenase iron protein 1 |
| 4e0v.1.A | 32.35 | homo-dimer | HHblits | X-ray | 3.10Å | 0.32 | 0.08 | L-amino-acid oxidase |
| 4e0v.1.B | 32.35 | homo-dimer | HHblits | X-ray | 3.10Å | 0.32 | 0.08 | L-amino-acid oxidase |
| 5ts5.1.A | 32.35 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | Amine oxidase |
| 5j1j.1.A | 29.41 | homo-dimer | HHblits | X-ray | 1.55Å | 0.32 | 0.08 | Site-determining protein |
| 5j1j.1.B | 29.41 | homo-dimer | HHblits | X-ray | 1.55Å | 0.32 | 0.08 | Site-determining protein |
| 5jvf.1.A | 29.41 | monomer | HHblits | X-ray | 1.66Å | 0.32 | 0.08 | Site-determining protein |
| 5gad.1.9 | 26.47 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.08 | Signal recognition particle receptor FtsY |
| 2y0d.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | UDP-GLUCOSE DEHYDROGENASE |
| 3enk.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | UDP-glucose 4-epimerase |
| 1npd.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB |
| 1npd.1.B | 13.89 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB |
| 1o9b.1.A | 13.89 | monomer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB |
| 1o9b.2.A | 13.89 | monomer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB |
| 1vi2.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | Shikimate 5-dehydrogenase 2 |
| 1vi2.1.B | 13.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | Shikimate 5-dehydrogenase 2 |
| 5swv.1.A | 13.89 | monomer | HHblits | X-ray | 2.65Å | 0.28 | 0.08 | Pentafunctional AROM polypeptide |
| 1ko5.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.28Å | 0.34 | 0.07 | Gluconate kinase |
| 1knq.1.B | 27.27 | homo-dimer | HHblits | X-ray | 2.00Å | 0.34 | 0.07 | Gluconate kinase |
| 1ko8.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.40Å | 0.34 | 0.07 | Gluconate kinase |
| 1ko8.1.B | 27.27 | homo-dimer | HHblits | X-ray | 2.40Å | 0.34 | 0.07 | Gluconate kinase |
| 1ko1.1.B | 27.27 | homo-dimer | HHblits | X-ray | 2.09Å | 0.34 | 0.07 | Gluconate kinase |
| 1knq.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.00Å | 0.34 | 0.07 | Gluconate kinase |
| 2xj4.1.A | 22.86 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | MIPZ |
| 5iaa.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 1.85Å | 0.30 | 0.08 | Ubiquitin-like modifier-activating enzyme 5 |
| 5iaa.1.B | 22.86 | hetero-oligomer | HHblits | X-ray | 1.85Å | 0.30 | 0.08 | Ubiquitin-like modifier-activating enzyme 5 |
| 3guc.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | Ubiquitin-like modifier-activating enzyme 5 |
| 3guc.1.B | 22.86 | homo-dimer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | Ubiquitin-like modifier-activating enzyme 5 |
| 2ffh.1.A | 22.86 | monomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | PROTEIN (FFH) |
| 1rj9.1.B | 22.86 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle protein |
| 2j7p.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 1.97Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 3ng1.1.A | 22.86 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | SIGNAL SEQUENCE RECOGNITION PROTEIN FFH |
| 2cnw.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.39Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2cnw.3.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.39Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2xkv.1.A | 22.86 | hetero-oligomer | HHblits | EM | 13.50Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c03.1.A | 22.86 | monomer | HHblits | X-ray | 1.24Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c03.2.A | 22.86 | monomer | HHblits | X-ray | 1.24Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1jpj.1.A | 22.86 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1jpn.1.A | 22.86 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1jpn.2.A | 22.86 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 3etd.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | GLUD1 protein |
| 3etg.1.D | 20.00 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Glutamate dehydrogenase |
| 1ls1.1.A | 22.86 | monomer | HHblits | X-ray | 1.10Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j45.1.A | 22.86 | monomer | HHblits | X-ray | 1.14Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j45.2.A | 22.86 | monomer | HHblits | X-ray | 1.14Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j46.1.A | 22.86 | monomer | HHblits | X-ray | 1.14Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2j46.2.A | 22.86 | monomer | HHblits | X-ray | 1.14Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1o87.1.A | 22.86 | monomer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 1o87.2.A | 22.86 | monomer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c04.1.A | 22.86 | monomer | HHblits | X-ray | 1.15Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2c04.2.A | 22.86 | monomer | HHblits | X-ray | 1.15Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE PROTEIN |
| 2ng1.1.A | 22.86 | monomer | HHblits | X-ray | 2.02Å | 0.30 | 0.08 | SIGNAL SEQUENCE RECOGNITION PROTEIN FFH |
| 3zmu.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A |
| 5h6q.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.53Å | 0.30 | 0.08 | Lysine-specific histone demethylase 1A |
| 2uxn.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.72Å | 0.30 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1 |
| 2dw4.1.A | 20.00 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Lysine-specific histone demethylase 1 |
| 2x0l.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1 |
| 3fwy.1.A | 22.86 | homo-dimer | HHblits | X-ray | 1.63Å | 0.30 | 0.08 | Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein |
| 3fwy.1.B | 22.86 | homo-dimer | HHblits | X-ray | 1.63Å | 0.30 | 0.08 | Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein |
| 2j37.1.G | 25.71 | hetero-oligomer | HHblits | EM | 8.00Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN (SRP54) |
| 4fos.1.A | 29.41 | monomer | HHblits | X-ray | 1.72Å | 0.32 | 0.08 | Shikimate dehydrogenase |
| 1rw4.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.50Å | 0.32 | 0.08 | Nitrogenase iron protein 1 |
| 1g20.1.E | 26.47 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.32 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g21.1.H | 26.47 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | NITROGENASE IRON PROTEIN |
| 1g21.1.E | 26.47 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | NITROGENASE IRON PROTEIN |
| 3cr7.1.A | 29.41 | homo-dimer | HHblits | X-ray | 2.50Å | 0.32 | 0.08 | Adenylyl-sulfate kinase |
| 3cr7.1.B | 29.41 | homo-dimer | HHblits | X-ray | 2.50Å | 0.32 | 0.08 | Adenylyl-sulfate kinase |
| 3uf0.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | Short-chain dehydrogenase/reductase SDR |
| 1uzm.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 1.49Å | 0.27 | 0.08 | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE |
| 1uzn.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 1.91Å | 0.27 | 0.08 | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE |
| 2ntn.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 3h7a.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 1.87Å | 0.27 | 0.08 | short chain dehydrogenase |
| 3dtt.1.A | 19.44 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.08 | NADP oxidoreductase |
| 5v2k.1.A | 22.86 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | UDP-glycosyltransferase 74F2 |
| 5v2k.2.A | 22.86 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | UDP-glycosyltransferase 74F2 |
| 3aoe.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | Glutamate dehydrogenase |
| 3aog.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Glutamate dehydrogenase |
| 3aog.1.D | 20.00 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | Glutamate dehydrogenase |
| 2wd7.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | PTERIDINE REDUCTASE |
| 4wcd.1.C | 20.00 | homo-tetramer | HHblits | X-ray | 1.68Å | 0.30 | 0.08 | Pteridine reductase |
| 1ffh.1.A | 22.86 | monomer | HHblits | X-ray | 2.05Å | 0.30 | 0.08 | FFH |
| 1ooe.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.65Å | 0.30 | 0.08 | Dihydropteridine reductase |
| 5l3s.1.B | 20.00 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3s.2.B | 20.00 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3s.3.B | 20.00 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3s.4.B | 20.00 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Signal recognition particle receptor FtsY |
| 5l3w.1.A | 20.00 | monomer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | Signal recognition particle receptor FtsY |
| 3u5t.1.A | 26.47 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.32 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 1ion.1.A | 26.47 | monomer | HHblits | X-ray | 2.30Å | 0.32 | 0.08 | PROBABLE CELL DIVISION INHIBITOR MIND |
| 1qzw.1.B | 23.53 | monomer | HHblits | X-ray | 4.10Å | 0.32 | 0.08 | Signal recognition 54 kDa protein |
| 1qzx.1.A | 23.53 | monomer | HHblits | X-ray | 4.00Å | 0.32 | 0.08 | Signal recognition 54 kDa protein |
| 3syn.2.A | 11.11 | hetero-oligomer | HHblits | X-ray | 3.06Å | 0.27 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.1.B | 11.11 | homo-dimer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.3.B | 11.11 | homo-dimer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.4.A | 11.11 | homo-dimer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px3.1.A | 11.11 | homo-dimer | HHblits | X-ray | 3.20Å | 0.27 | 0.08 | Flagellar biosynthesis protein flhF |
| 2px0.1.A | 11.11 | homo-dimer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | Flagellar biosynthesis protein flhF |
| 1bg6.1.A | 30.30 | homo-dimer | HHblits | X-ray | 1.80Å | 0.34 | 0.07 | N-(1-D-CARBOXYLETHYL)-L-NORVALINE DEHYDROGENASE |
| 1x1e.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.76Å | 0.29 | 0.08 | 2-deoxy-D-gluconate 3-dehydrogenase |
| 4n5l.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.29 | 0.08 | Acetoacetyl-CoA reductase |
| 4n5m.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 1.34Å | 0.29 | 0.08 | Acetoacetyl-CoA reductase |
| 1l1f.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | Glutamate Dehydrogenase 1 |
| 3ctm.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.29 | 0.08 | Carbonyl Reductase |
| 3ctm.1.B | 20.00 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.29 | 0.08 | Carbonyl Reductase |
| 3ctm.1.C | 20.00 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.29 | 0.08 | Carbonyl Reductase |
| 3ctm.1.D | 20.00 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.29 | 0.08 | Carbonyl Reductase |
| 3ctm.2.B | 20.00 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.29 | 0.08 | Carbonyl Reductase |
| 3ctm.2.C | 20.00 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.29 | 0.08 | Carbonyl Reductase |
| 2k7f.1.A | 14.29 | monomer | HHblits | NMR | NA | 0.29 | 0.08 | Replication factor C subunit 1 |
| 2k6g.1.A | 14.29 | monomer | HHblits | NMR | NA | 0.29 | 0.08 | Replication factor C subunit 1 |
| 5vj7.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.55Å | 0.29 | 0.08 | Oxidoreductase |
| 5jca.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.29 | 0.08 | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) subunit alpha |
| 2hae.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 3.13Å | 0.29 | 0.08 | Malate oxidoreductase |
| 1vl6.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.61Å | 0.29 | 0.08 | malate oxidoreductase |
| 1vl6.1.B | 22.86 | homo-tetramer | HHblits | X-ray | 2.61Å | 0.29 | 0.08 | malate oxidoreductase |
| 3dm9.1.A | 22.86 | homo-hexamer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Signal recognition particle receptor |
| 3e70.1.A | 22.86 | monomer | HHblits | X-ray | 1.97Å | 0.29 | 0.08 | Signal recognition particle receptor |
| 3dmd.1.F | 22.86 | homo-hexamer | HHblits | X-ray | 2.21Å | 0.29 | 0.08 | Signal recognition particle receptor |
| 3dmd.1.E | 22.86 | homo-hexamer | HHblits | X-ray | 2.21Å | 0.29 | 0.08 | Signal recognition particle receptor |
| 1ww8.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | malate oxidoreductase |
| 4b3h.1.A | 22.86 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | FATTY ACID BETA-OXIDATION COMPLEX ALPHA-CHAIN FADB |
| 4b3h.1.B | 22.86 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | FATTY ACID BETA-OXIDATION COMPLEX ALPHA-CHAIN FADB |
| 4ylf.1.B | 20.00 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | Dihydropyrimidine dehydrogenase subunit A |
| 4ylf.2.B | 20.00 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | Dihydropyrimidine dehydrogenase subunit A |
| 4yry.1.B | 20.00 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | Dihydropyrimidine dehydrogenase subunit A |
| 4yry.2.B | 20.00 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | Dihydropyrimidine dehydrogenase subunit A |
| 1tdo.1.A | 32.35 | monomer | HHblits | X-ray | 3.00Å | 0.32 | 0.08 | L-amino acid oxidase |
| 2y0c.1.A | 13.89 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.08 | UDP-GLUCOSE DEHYDROGENASE |
| 2y0c.2.A | 13.89 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.08 | UDP-GLUCOSE DEHYDROGENASE |
| 2y0e.1.A | 13.89 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.08 | UDP-GLUCOSE DEHYDROGENASE |
| 2y0e.2.A | 13.89 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.08 | UDP-GLUCOSE DEHYDROGENASE |
| 2egg.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase |
| 2xox.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | PTERIDINE REDUCTASE |
| 2xox.1.B | 19.44 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | PTERIDINE REDUCTASE |
| 3pwz.1.A | 13.89 | monomer | HHblits | X-ray | 1.71Å | 0.27 | 0.08 | Shikimate dehydrogenase 3 |
| 4usq.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.39Å | 0.27 | 0.08 | PYRIDINE NUCLEOTIDE-DISULFIDE OXIDOREDUCTASE |
| 4cqm.1.B | 13.89 | hetero-oligomer | HHblits | X-ray | 2.34Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.1.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.2.B | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.2.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.3.B | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.3.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.4.B | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.4.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cql.1.B | 13.89 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cqm.2.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.34Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cqm.3.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.34Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 4cqm.4.C | 13.89 | hetero-oligomer | HHblits | X-ray | 2.34Å | 0.27 | 0.08 | CARBONYL REDUCTASE FAMILY MEMBER 4 |
| 1l7b.1.A | 11.43 | monomer | HHblits | NMR | NA | 0.29 | 0.08 | DNA LIGASE |
| 5u6m.1.A | 22.86 | monomer | HHblits | X-ray | 2.57Å | 0.29 | 0.08 | UDP-glycosyltransferase 74F2 |
| 5u6m.2.A | 22.86 | monomer | HHblits | X-ray | 2.57Å | 0.29 | 0.08 | UDP-glycosyltransferase 74F2 |
| 5u6s.1.A | 22.86 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | UDP-glycosyltransferase 74F2 |
| 5u6s.2.A | 22.86 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | UDP-glycosyltransferase 74F2 |
| 3dm5.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.51Å | 0.29 | 0.08 | Signal recognition 54 kDa protein |
| 4p22.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.08 | Ubiquitin-like modifier-activating enzyme 1 |
| 4p22.1.B | 22.86 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.08 | Ubiquitin-like modifier-activating enzyme 1 |
| 5ff5.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.93Å | 0.29 | 0.08 | PaaA |
| 5ff5.1.B | 22.86 | homo-dimer | HHblits | X-ray | 2.93Å | 0.29 | 0.08 | PaaA |
| 4y4n.1.A | 20.00 | homo-octamer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | Putative ribose 1,5-bisphosphate isomerase |
| 2qsy.1.A | 23.53 | monomer | HHblits | X-ray | 1.95Å | 0.31 | 0.08 | Nicotinamide riboside kinase 1 |
| 2qt0.1.A | 23.53 | monomer | HHblits | X-ray | 1.92Å | 0.31 | 0.08 | Nicotinamide riboside kinase 1 |
| 3h5n.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | MccB protein |
| 3h5n.1.C | 20.59 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | MccB protein |
| 3h9q.2.A | 20.59 | homo-dimer | HHblits | X-ray | 2.63Å | 0.31 | 0.08 | MccB protein |
| 5u1j.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.95Å | 0.31 | 0.08 | Uncharacterized protein |
| 5u1j.1.B | 26.47 | homo-dimer | HHblits | X-ray | 2.95Å | 0.31 | 0.08 | Uncharacterized protein |
| 5u1j.2.A | 26.47 | homo-dimer | HHblits | X-ray | 2.95Å | 0.31 | 0.08 | Uncharacterized protein |
| 5u1j.2.B | 26.47 | homo-dimer | HHblits | X-ray | 2.95Å | 0.31 | 0.08 | Uncharacterized protein |
| 3iqx.1.A | 29.41 | homo-dimer | HHblits | X-ray | 3.50Å | 0.31 | 0.08 | Tail-anchored protein targeting factor Get3 |
| 4v03.1.A | 26.47 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | SITE-DETERMINING PROTEIN |
| 3kve.1.A | 32.35 | homo-tetramer | HHblits | X-ray | 2.57Å | 0.31 | 0.08 | L-amino acid oxidase |
| 3adb.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | L-seryl-tRNA(Sec) kinase |
| 3adc.1.B | 26.47 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.08 | L-seryl-tRNA(Sec) kinase |
| 3adc.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.08 | L-seryl-tRNA(Sec) kinase |
| 3adb.1.B | 26.47 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.08 | L-seryl-tRNA(Sec) kinase |
| 3add.1.B | 26.47 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.08 | L-seryl-tRNA(Sec) kinase |
| 5gad.1.9 | 26.47 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.08 | Signal recognition particle receptor FtsY |
| 2yhs.1.A | 26.47 | monomer | HHblits | X-ray | 1.60Å | 0.31 | 0.08 | CELL DIVISION PROTEIN FTSY |
| 5gud.1.A | 19.44 | homo-hexamer | HHblits | X-ray | 1.68Å | 0.27 | 0.08 | Glutamate dehydrogenase |
| 5gud.1.D | 19.44 | homo-hexamer | HHblits | X-ray | 1.68Å | 0.27 | 0.08 | Glutamate dehydrogenase |
| 4xdb.1.A | 11.11 | homo-dimer | HHblits | X-ray | 3.32Å | 0.27 | 0.08 | NADH dehydrogenase-like protein SAOUHSC_00878 |
| 3o26.1.A | 13.89 | monomer | HHblits | X-ray | 1.91Å | 0.27 | 0.08 | Salutaridine reductase |
| 2xj4.1.A | 22.22 | monomer | HHblits | X-ray | 1.60Å | 0.27 | 0.08 | MIPZ |
| 2xit.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | MIPZ |
| 4pcv.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.05Å | 0.29 | 0.08 | BdcA (YjgI) |
| 3icc.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 1.87Å | 0.29 | 0.08 | Putative 3-oxoacyl-(acyl carrier protein) reductase |
| 3mog.1.A | 22.86 | homo-trimer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Probable 3-hydroxybutyryl-CoA dehydrogenase |
| 3mog.1.B | 22.86 | homo-trimer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | Probable 3-hydroxybutyryl-CoA dehydrogenase |
| 5u2w.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.29 | 0.08 | Short chain dehydrogenase |
| 3d1c.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | Flavin-containing Putative Monooxygenase |
| 2xlr.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.55Å | 0.29 | 0.08 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xlp.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xlt.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xls.1.A | 17.14 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.08 | FLAVIN-CONTAINING MONOOXYGENASE |
| 3dm5.1.A | 30.30 | homo-tetramer | HHblits | X-ray | 2.51Å | 0.34 | 0.07 | Signal recognition 54 kDa protein |
| 1t2a.1.A | 29.41 | homo-tetramer | HHblits | X-ray | 1.84Å | 0.31 | 0.08 | GDP-mannose 4,6 dehydratase |
| 1g3r.1.A | 26.47 | monomer | HHblits | X-ray | 2.70Å | 0.31 | 0.08 | CELL DIVISION INHIBITOR |
| 4h2r.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.47Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 4h2r.1.D | 17.65 | homo-tetramer | HHblits | X-ray | 2.47Å | 0.31 | 0.08 | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase |
| 4w7h.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 3.11Å | 0.31 | 0.08 | Carbonyl reductase |
| 3fbt.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.08 | chorismate mutase and shikimate 5-dehydrogenase fusion protein |
| 1p77.1.A | 16.67 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase |
| 1p74.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase |
| 1p74.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase |
| 4ndn.1.E | 11.11 | hetero-oligomer | HHblits | X-ray | 2.34Å | 0.27 | 0.08 | Methionine adenosyltransferase 2 subunit beta |
| 4ndn.1.F | 11.11 | hetero-oligomer | HHblits | X-ray | 2.34Å | 0.27 | 0.08 | Methionine adenosyltransferase 2 subunit beta |
| 4gvl.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | TrkA domain protein |
| 4gvl.1.B | 16.67 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | TrkA domain protein |
| 4gvl.1.C | 16.67 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | TrkA domain protein |
| 4gvl.1.D | 16.67 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | TrkA domain protein |
| 6aqy.1.A | 8.11 | homo-dimer | HHblits | X-ray | 2.55Å | 0.25 | 0.08 | gdp-l-fucose synthetase |
| 6aqz.2.A | 8.11 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.08 | gdp-l-fucose synthetase |
| 1nr1.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Glutamate dehydrogenase 1 |
| 1nr1.1.C | 17.14 | homo-hexamer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Glutamate dehydrogenase 1 |
| 1nqt.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 3.50Å | 0.29 | 0.08 | Glutamate dehydrogenase 1 |
| 1nqt.1.C | 17.14 | homo-hexamer | HHblits | X-ray | 3.50Å | 0.29 | 0.08 | Glutamate dehydrogenase 1 |
| 1nqt.1.E | 17.14 | homo-hexamer | HHblits | X-ray | 3.50Å | 0.29 | 0.08 | Glutamate dehydrogenase 1 |
| 3r3j.1.A | 28.57 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.29 | 0.08 | Glutamate dehydrogenase |
| 3l77.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.29 | 0.08 | Short-chain alcohol dehydrogenase |
| 2bac.1.A | 11.43 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | putative aminooxidase |
| 4fda.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| 2vdc.1.G | 20.00 | hetero-oligomer | HHblits | EM | 9.50Å | 0.29 | 0.08 | GLUTAMATE SYNTHASE [NADPH] SMALL CHAIN |
| 3of5.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.52Å | 0.29 | 0.08 | Dethiobiotin synthetase |
| 3nwj.1.A | 21.21 | monomer | HHblits | X-ray | 2.35Å | 0.33 | 0.07 | AtSK2 |
| 3nwj.2.A | 21.21 | monomer | HHblits | X-ray | 2.35Å | 0.33 | 0.07 | AtSK2 |
| 3zyx.1.A | 36.36 | homo-dimer | HHblits | X-ray | 2.20Å | 0.33 | 0.07 | AMINE OXIDASE [FLAVIN-CONTAINING] B |
| 2xfu.1.A | 36.36 | homo-dimer | HHblits | X-ray | 2.20Å | 0.33 | 0.07 | AMINE OXIDASE [FLAVIN-CONTAINING] B |
| 3ibg.1.A | 36.36 | homo-dimer | HHblits | X-ray | 3.20Å | 0.33 | 0.07 | ATPase, subunit of the Get complex |
| 3gwf.1.A | 26.47 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.08 | Cyclohexanone monooxygenase |
| 3gwd.1.A | 26.47 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.08 | Cyclohexanone monooxygenase |
| 3b9q.1.A | 23.53 | monomer | HHblits | X-ray | 1.75Å | 0.31 | 0.08 | Chloroplast SRP receptor homolog, alpha subunit CPFTSY |
| 5nco.1.9 | 20.59 | hetero-oligomer | HHblits | EM | 4.80Å | 0.31 | 0.08 | Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein,Signal recognition particle protein |
| 1f8s.1.A | 29.41 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | L-AMINO ACID OXIDASE |
| 2ylr.1.A | 11.11 | monomer | HHblits | X-ray | 2.26Å | 0.26 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 1w4x.1.A | 11.11 | monomer | HHblits | X-ray | 1.70Å | 0.26 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 4d03.1.A | 11.11 | monomer | HHblits | X-ray | 1.81Å | 0.26 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 5tgd.1.A | 16.22 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.24 | 0.08 | FolM Alternative dihydrofolate reductase |
| 4omu.1.A | 10.81 | monomer | HHblits | X-ray | 1.65Å | 0.24 | 0.08 | Shikimate dehydrogenase |
| 4omu.2.A | 10.81 | monomer | HHblits | X-ray | 1.65Å | 0.24 | 0.08 | Shikimate dehydrogenase |
| 3kyd.1.B | 22.86 | hetero-oligomer | HHblits | X-ray | 2.61Å | 0.29 | 0.08 | SUMO-activating enzyme subunit 2 |
| 1y8q.1.B | 22.86 | hetero-oligomer | HHblits | X-ray | 2.25Å | 0.29 | 0.08 | Ubiquitin-like 2 activating enzyme E1B |
| 1y8q.2.B | 22.86 | hetero-oligomer | HHblits | X-ray | 2.25Å | 0.29 | 0.08 | Ubiquitin-like 2 activating enzyme E1B |
| 1y8r.1.B | 22.86 | hetero-oligomer | HHblits | X-ray | 2.75Å | 0.29 | 0.08 | Ubiquitin-like 2 activating enzyme E1B |
| 1y8r.2.B | 22.86 | hetero-oligomer | HHblits | X-ray | 2.75Å | 0.29 | 0.08 | Ubiquitin-like 2 activating enzyme E1B |
| 3edm.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | Short chain dehydrogenase |
| 3edm.1.C | 20.00 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | Short chain dehydrogenase |
| 4xij.1.A | 14.29 | monomer | HHblits | X-ray | 1.45Å | 0.29 | 0.08 | Shikimate 5-dehydrogenase |
| 2gqf.1.A | 22.86 | monomer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | Hypothetical protein HI0933 |
| 2jb2.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.45Å | 0.29 | 0.08 | L-AMINO ACID OXIDASE |
| 1zu5.1.A | 11.43 | monomer | HHblits | X-ray | 2.40Å | 0.29 | 0.08 | ftsY |
| 1zu4.1.A | 11.43 | monomer | HHblits | X-ray | 1.95Å | 0.29 | 0.08 | ftsY |
| 2rgj.1.A | 26.47 | monomer | HHblits | X-ray | 2.40Å | 0.31 | 0.08 | Flavin-containing monooxygenase |
| 3c96.1.A | 26.47 | monomer | HHblits | X-ray | 1.90Å | 0.31 | 0.08 | Flavin-containing monooxygenase |
| 4j2w.2.A | 23.53 | monomer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Kynurenine 3-monooxygenase |
| 4j2w.1.A | 23.53 | monomer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Kynurenine 3-monooxygenase |
| 4j33.1.A | 23.53 | monomer | HHblits | X-ray | 1.82Å | 0.31 | 0.08 | Kynurenine 3-monooxygenase |
| 4j33.2.A | 23.53 | monomer | HHblits | X-ray | 1.82Å | 0.31 | 0.08 | Kynurenine 3-monooxygenase |
| 4j36.1.A | 23.53 | monomer | HHblits | X-ray | 2.13Å | 0.31 | 0.08 | Kynurenine 3-monooxygenase |
| 4j36.2.A | 23.53 | monomer | HHblits | X-ray | 2.13Å | 0.31 | 0.08 | Kynurenine 3-monooxygenase |
| 1j8y.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | SIGNAL RECOGNITION 54 KDA PROTEIN |
| 4y4l.1.A | 26.47 | homo-octamer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | Thiamine thiazole synthase |
| 5l3r.1.A | 23.53 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.31 | 0.08 | Signal recognition particle 54 kDa protein, chloroplastic |
| 3ibg.1.A | 26.47 | homo-dimer | HHblits | X-ray | 3.20Å | 0.31 | 0.08 | ATPase, subunit of the Get complex |
| 4kwh.1.A | 19.44 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.26 | 0.08 | Reductase homolog |
| 5mh4.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.14Å | 0.28 | 0.08 | Thioredoxin reductase |
| 5mjk.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | Thioredoxin reductase |
| 3x0v.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | L-lysine oxidase |
| 1nvt.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.08 | Shikimate 5'-dehydrogenase |
| 1nvt.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.08 | Shikimate 5'-dehydrogenase |
| 3md0.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.45Å | 0.28 | 0.08 | Arginine/ornithine transport system ATPase |
| 2ynm.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | LIGHT-INDEPENDENT PROTOCHLOROPHYLLIDE REDUCTASE IRON-SULFUR ATP-BINDING PROTEIN |
| 1cp2.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.93Å | 0.31 | 0.08 | NITROGENASE IRON PROTEIN |
| 1cp2.1.B | 23.53 | homo-dimer | HHblits | X-ray | 1.93Å | 0.31 | 0.08 | NITROGENASE IRON PROTEIN |
| 4lw8.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.08 | Putative epimerase |
| 2gah.1.B | 26.47 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.31 | 0.08 | heterotetrameric sarcosine oxidase beta-subunit |
| 1hyq.1.A | 23.53 | monomer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | CELL DIVISION INHIBITOR (MIND-1) |
| 4wop.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.39Å | 0.31 | 0.08 | ATP-dependent dethiobiotin synthetase BioD |
| 3r9i.1.B | 23.53 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.31 | 0.08 | Septum site-determining protein minD |
| 3q9l.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.34Å | 0.31 | 0.08 | Septum site-determining protein minD |
| 3o8q.1.B | 11.11 | hetero-oligomer | HHblits | X-ray | 1.45Å | 0.26 | 0.08 | Shikimate 5-dehydrogenase I alpha |
| 3o8q.1.A | 11.11 | hetero-oligomer | HHblits | X-ray | 1.45Å | 0.26 | 0.08 | Shikimate 5-dehydrogenase I alpha |
| 3sef.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.08 | Shikimate 5-dehydrogenase |
| 3sef.1.B | 11.11 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.08 | Shikimate 5-dehydrogenase |
| 3sef.2.A | 11.11 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.08 | Shikimate 5-dehydrogenase |
| 3sef.2.B | 11.11 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.08 | Shikimate 5-dehydrogenase |
| 3tnl.1.B | 11.11 | homo-dimer | HHblits | X-ray | 1.45Å | 0.26 | 0.08 | Shikimate dehydrogenase |
| 3tnl.1.A | 11.11 | homo-dimer | HHblits | X-ray | 1.45Å | 0.26 | 0.08 | Shikimate dehydrogenase |
| 3toz.1.B | 11.11 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.08 | Shikimate dehydrogenase |
| 3toz.3.A | 11.11 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.08 | Shikimate dehydrogenase |
| 3don.1.A | 11.11 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | Shikimate dehydrogenase |
| 3ksu.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.08 | 3-oxoacyl-acyl carrier protein reductase |
| 4cjx.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.08 | C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, PUTATIVE |
| 4cjx.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.08 | C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, PUTATIVE |
| 1t3h.1.A | 34.38 | monomer | HHblits | X-ray | 2.50Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1t3h.2.A | 34.38 | monomer | HHblits | X-ray | 2.50Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1t3h.3.A | 34.38 | monomer | HHblits | X-ray | 2.50Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1vhl.1.A | 34.38 | monomer | HHblits | X-ray | 1.65Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1vhl.2.A | 34.38 | monomer | HHblits | X-ray | 1.65Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1vhl.3.A | 34.38 | monomer | HHblits | X-ray | 1.65Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1viy.1.A | 34.38 | homo-trimer | HHblits | X-ray | 1.89Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1viy.1.B | 34.38 | homo-trimer | HHblits | X-ray | 1.89Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1viy.1.C | 34.38 | homo-trimer | HHblits | X-ray | 1.89Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1n3b.1.A | 34.38 | homo-trimer | HHblits | X-ray | 1.80Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1n3b.1.B | 34.38 | homo-trimer | HHblits | X-ray | 1.80Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 1n3b.1.C | 34.38 | homo-trimer | HHblits | X-ray | 1.80Å | 0.35 | 0.07 | Dephospho-CoA kinase |
| 3e9n.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE |
| 2qhx.1.A | 22.86 | homo-tetramer | HHblits | X-ray | 2.61Å | 0.28 | 0.08 | Pteridine reductase 1 |
| 3jd2.1.B | 17.14 | homo-hexamer | HHblits | EM | NA | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 3mvo.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 3.23Å | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 3qmu.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 3.62Å | 0.28 | 0.08 | Glutamate dehydrogenase 1 |
| 3mw9.1.E | 17.14 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | Glutamate dehydrogenase 1 |
| 3mvo.1.B | 17.14 | homo-hexamer | HHblits | X-ray | 3.23Å | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 3qmu.1.B | 17.14 | homo-hexamer | HHblits | X-ray | 3.62Å | 0.28 | 0.08 | Glutamate dehydrogenase 1 |
| 3jd0.1.B | 17.14 | homo-hexamer | HHblits | EM | NA | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 3jd3.1.A | 17.14 | homo-hexamer | HHblits | EM | NA | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 3jd4.1.B | 17.14 | homo-hexamer | HHblits | EM | NA | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 3jd1.1.C | 17.14 | homo-hexamer | HHblits | EM | NA | 0.28 | 0.08 | Glutamate dehydrogenase 1, mitochondrial |
| 5l6h.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5l6i.2.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.76Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4nnj.1.A | 20.00 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4nnj.1.C | 20.00 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5jyd.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.28 | 0.08 | Short chain dehydrogenase |
| 3ug7.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.28 | 0.08 | arsenical pump-driving ATPase |
| 3ug6.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 3.30Å | 0.28 | 0.08 | arsenical pump-driving ATPase |
| 1mfz.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | GDP-mannose 6-dehydrogenase |
| 3aus.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | Glucose 1-dehydrogenase 4 |
| 1djn.1.A | 33.33 | homo-dimer | HHblits | X-ray | 2.20Å | 0.33 | 0.07 | TRIMETHYLAMINE DEHYDROGENASE |
| 1djq.1.A | 33.33 | homo-dimer | HHblits | X-ray | 2.20Å | 0.33 | 0.07 | TRIMETHYLAMINE DEHYDROGENASE |
| 3n2i.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.25Å | 0.33 | 0.07 | Thymidylate kinase |
| 4ffm.1.A | 26.47 | monomer | HHblits | X-ray | 1.91Å | 0.30 | 0.08 | PylC |
| 2c8v.1.A | 26.47 | monomer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN 1 |
| 1de0.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | NITROGENASE IRON PROTEIN |
| 1xcp.1.A | 26.47 | homo-dimer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 1xcp.2.A | 26.47 | homo-dimer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 4wza.1.E | 26.47 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 4wza.1.F | 26.47 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 4wzb.1.F | 26.47 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 4wzb.1.E | 26.47 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 3v76.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.51Å | 0.30 | 0.08 | Flavoprotein |
| 4lis.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.08 | UDP-glucose 4-epimerase |
| 2ylz.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 2ylx.1.A | 11.11 | monomer | HHblits | X-ray | 2.20Å | 0.26 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 2ym2.1.A | 11.11 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 3tdk.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 4iiu.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5aun.1.B | 25.71 | hetero-oligomer | HHblits | X-ray | 1.63Å | 0.28 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 3vx3.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 5auq.3.A | 25.71 | homo-dimer | HHblits | X-ray | 2.53Å | 0.28 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 3vx3.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog |
| 4ohv.1.A | 17.14 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | Protein clpf-1 |
| 4eit.1.A | 11.11 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.26 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5tf4.1.A | 11.11 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.26 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 5tf4.2.B | 11.11 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.26 | 0.08 | Enoyl-[acyl-carrier-protein] reductase [NADH] |
| 3uov.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.08 | OTEMO |
| 3uoz.1.A | 11.11 | monomer | HHblits | X-ray | 2.41Å | 0.26 | 0.08 | OTEMO |
| 3up4.1.A | 11.11 | monomer | HHblits | X-ray | 2.80Å | 0.26 | 0.08 | OTEMO |
| 1kyq.2.A | 13.89 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.08 | Siroheme biosynthesis protein MET8 |
| 1kyq.1.B | 13.89 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.08 | Siroheme biosynthesis protein MET8 |
| 1kyq.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.08 | Siroheme biosynthesis protein MET8 |
| 2ph3.1.A | 23.53 | homo-tetramer | HHblits | X-ray | 1.91Å | 0.30 | 0.08 | 3-oxoacyl-[acyl carrier protein] reductase |
| 2ph3.1.B | 23.53 | homo-tetramer | HHblits | X-ray | 1.91Å | 0.30 | 0.08 | 3-oxoacyl-[acyl carrier protein] reductase |
| 4da9.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Short-chain dehydrogenase/reductase |
| 4da9.1.B | 17.65 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Short-chain dehydrogenase/reductase |
| 4da9.1.C | 17.65 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Short-chain dehydrogenase/reductase |
| 4da9.1.D | 17.65 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | Short-chain dehydrogenase/reductase |
| 3k9h.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | PF-32 protein |
| 3k9g.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.25Å | 0.30 | 0.08 | PF-32 protein |
| 1d4c.2.A | 20.59 | monomer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | FLAVOCYTOCHROME C FUMARATE REDUCTASE |
| 1d4c.1.A | 20.59 | monomer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | FLAVOCYTOCHROME C FUMARATE REDUCTASE |
| 1d4d.1.A | 20.59 | monomer | HHblits | X-ray | 2.50Å | 0.30 | 0.08 | FLAVOCYTOCHROME C FUMARATE REDUCTASE |
| 1wdm.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 3.80Å | 0.30 | 0.08 | Fatty oxidation complex alpha subunit |
| 1wdl.1.B | 20.59 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.08 | Fatty oxidation complex alpha subunit |
| 1wdl.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.08 | Fatty oxidation complex alpha subunit |
| 2d3t.1.B | 20.59 | hetero-oligomer | HHblits | X-ray | 3.40Å | 0.30 | 0.08 | Fatty oxidation complex alpha subunit |
| 1jrz.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | FLAVOCYTOCHROME C |
| 1e39.1.A | 26.47 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | FUMARATE REDUCTASE FLAVOPROTEIN SUBUNIT |
| 2b7s.1.A | 26.47 | monomer | HHblits | X-ray | 2.12Å | 0.30 | 0.08 | Fumarate reductase flavoprotein subunit |
| 1m64.1.A | 26.47 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | flavocytochrome c3 |
| 1lj1.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | flavocytochrome c3 |
| 1kss.1.A | 26.47 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | flavocytochrome c |
| 1jrx.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | FLAVOCYTOCHROME C |
| 1jry.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | FLAVOCYTOCHROME C |
| 1ksu.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | flavocytochrome c |
| 3ado.1.A | 29.41 | homo-dimer | HHblits | X-ray | 1.70Å | 0.30 | 0.08 | Lambda-crystallin |
| 4k8d.1.A | 26.47 | homo-dimer | HHblits | X-ray | 1.86Å | 0.30 | 0.08 | Mercuric reductase |
| 3kpf.1.A | 26.47 | monomer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Polyamine oxidase |
| 3ku9.2.A | 26.47 | monomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Polyamine oxidase |
| 1h81.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.08 | POLYAMINE OXIDASE |
| 4fq8.1.A | 30.30 | monomer | HHblits | X-ray | 2.07Å | 0.33 | 0.07 | Shikimate dehydrogenase |
| 4fq8.2.A | 30.30 | monomer | HHblits | X-ray | 2.07Å | 0.33 | 0.07 | Shikimate dehydrogenase |
| 5um6.1.A | 14.29 | monomer | HHblits | X-ray | 2.79Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4ii3.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 4ii2.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5knl.2.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 5knl.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 2j3h.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3h.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3k.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3k.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3j.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3j.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3i.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 2j3i.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.80Å | 0.28 | 0.08 | NADP-DEPENDENT OXIDOREDUCTASE P1 |
| 3klj.1.A | 17.14 | monomer | HHblits | X-ray | 2.10Å | 0.28 | 0.08 | NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) |
| 3vps.1.A | 22.22 | monomer | HHblits | X-ray | 1.90Å | 0.26 | 0.08 | NAD-dependent epimerase/dehydratase |
| 1xd9.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | Nitrogenase iron protein 1 |
| 3ada.1.B | 26.47 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | SARCOSINE OXIDASE BETA SUBUNIT |
| 3zq6.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.11Å | 0.30 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 3zq6.1.B | 17.65 | homo-dimer | HHblits | X-ray | 2.11Å | 0.30 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 3zq6.2.A | 17.65 | homo-dimer | HHblits | X-ray | 2.11Å | 0.30 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 3zq6.2.B | 17.65 | homo-dimer | HHblits | X-ray | 2.11Å | 0.30 | 0.08 | PUTATIVE ARSENICAL PUMP-DRIVING ATPASE |
| 4ue5.1.D | 26.47 | hetero-oligomer | HHblits | EM | 9.00Å | 0.30 | 0.08 | SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN |
| 1fk8.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.95Å | 0.32 | 0.07 | 3ALPHA-HYDROXYSTEROID DEHYDROGENASE/CARBONYL REDUCTASE |
| 1fk8.1.B | 27.27 | homo-dimer | HHblits | X-ray | 1.95Å | 0.32 | 0.07 | 3ALPHA-HYDROXYSTEROID DEHYDROGENASE/CARBONYL REDUCTASE |
| 3a28.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.32 | 0.07 | L-2.3-butanediol dehydrogenase |
| 5u63.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.99Å | 0.28 | 0.08 | Thioredoxin reductase |
| 4osp.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.28 | 0.08 | Oxygenase-reductase |
| 1uzl.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE |
| 3hdq.1.A | 25.71 | homo-dimer | HHblits | X-ray | 2.36Å | 0.28 | 0.08 | UDP-galactopyranose mutase |
| 3mj4.1.B | 25.71 | homo-dimer | HHblits | X-ray | 2.65Å | 0.28 | 0.08 | UDP-galactopyranose mutase |
| 3mj4.2.A | 25.71 | homo-dimer | HHblits | X-ray | 2.65Å | 0.28 | 0.08 | UDP-galactopyranose mutase |
| 5vt3.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.98Å | 0.28 | 0.08 | Thioredoxin reductase |
| 5eqf.1.A | 19.44 | homo-dimer | HHblits | X-ray | 2.14Å | 0.25 | 0.08 | UDP-galactopyranose mutase |
| 1kkl.1.A | 26.47 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | HprK protein |
| 1kkl.1.B | 26.47 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | HprK protein |
| 1jb1.1.A | 26.47 | homo-hexamer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | HPRK PROTEIN |
| 2qmh.1.B | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.1.A | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.1.C | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.1.E | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.1.F | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.2.A | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.2.C | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.2.E | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 2qmh.2.F | 26.47 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.30 | 0.08 | HPr kinase/phosphorylase |
| 3vzr.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Acetoacetyl-CoA reductase |
| 2a8x.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | Dihydrolipoyl dehydrogenase |
| 3djd.1.A | 29.41 | monomer | HHblits | X-ray | 1.75Å | 0.30 | 0.08 | Fructosyl amine: oxygen oxidoreductase |
| 3dje.1.A | 29.41 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.08 | Fructosyl amine: oxygen oxidoreductase |
| 4a9w.1.A | 26.47 | homo-dimer | HHblits | X-ray | 2.72Å | 0.30 | 0.08 | MONOOXYGENASE |
| 3tn7.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 1.68Å | 0.30 | 0.08 | Short-chain alcohol dehydrogenase |
| 4kug.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4kug.2.B | 20.59 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4kue.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4kue.1.B | 20.59 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4kuh.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.51Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4kuh.1.B | 20.59 | homo-dimer | HHblits | X-ray | 2.51Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4kuh.3.B | 20.59 | homo-dimer | HHblits | X-ray | 2.51Å | 0.30 | 0.08 | 3-hydroxybutyryl-CoA dehydrogenase |
| 1ps9.1.A | 23.53 | monomer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | 2,4-dienoyl-CoA reductase |
| 5u1g.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 3.64Å | 0.30 | 0.08 | ParA |
| 5u1g.2.B | 20.59 | hetero-oligomer | HHblits | X-ray | 3.64Å | 0.30 | 0.08 | ParA |
| 4e07.1.A | 20.59 | monomer | HHblits | X-ray | 2.90Å | 0.30 | 0.08 | Plasmid partitioning protein ParF |
| 4e09.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.99Å | 0.30 | 0.08 | Plasmid partitioning protein ParF |
| 4e03.1.A | 20.59 | monomer | HHblits | X-ray | 2.45Å | 0.30 | 0.08 | Plasmid partitioning protein ParF |
| 4e03.2.A | 20.59 | monomer | HHblits | X-ray | 2.45Å | 0.30 | 0.08 | Plasmid partitioning protein ParF |
| 4dzz.1.A | 20.59 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | Plasmid partitioning protein ParF |
| 4dzz.2.A | 20.59 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | Plasmid partitioning protein ParF |
| 4pfs.1.A | 23.53 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Cobyrinic Acid a,c-diamide synthase |
| 4pfs.2.A | 23.53 | monomer | HHblits | X-ray | 2.30Å | 0.30 | 0.08 | Cobyrinic Acid a,c-diamide synthase |
| 5if9.1.A | 23.53 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.08 | Cobyrinic Acid a,c-diamide synthase |
| 5l3q.1.A | 26.47 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Signal recognition particle 54 kDa protein |
| 1zzg.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.95Å | 0.32 | 0.07 | glucose-6-phosphate isomerase |
| 4xgi.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | Glutamate dehydrogenase |
| 4u8j.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 3afn.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 1.63Å | 0.27 | 0.08 | Carbonyl reductase |
| 3afm.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.27 | 0.08 | Carbonyl reductase |
| 5cee.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | NAD-dependent malic enzyme |
| 4gx5.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 3.70Å | 0.27 | 0.08 | TrkA domain protein |
| 1ki9.1.A | 28.13 | homo-trimer | HHblits | X-ray | 2.76Å | 0.34 | 0.07 | adenylate kinase |
| 1ki9.1.B | 28.13 | homo-trimer | HHblits | X-ray | 2.76Å | 0.34 | 0.07 | adenylate kinase |
| 1ki9.1.C | 28.13 | homo-trimer | HHblits | X-ray | 2.76Å | 0.34 | 0.07 | adenylate kinase |
| 4wct.1.A | 26.47 | monomer | HHblits | X-ray | 1.67Å | 0.30 | 0.08 | Fructosyl amine:oxygen oxidoreductase |
| 2y48.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.30 | 0.08 | LYSINE-SPECIFIC DEMETHYLASE 1A |
| 3abu.1.A | 20.59 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.08 | Lysine-specific histone demethylase 1 |
| 4bay.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.08 | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A |
| 5lgn.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.08 | Lysine-specific histone demethylase 1A |
| 4y4m.1.A | 23.53 | homo-octamer | HHblits | X-ray | 2.71Å | 0.30 | 0.08 | Putative ribose 1,5-bisphosphate isomerase |
| 5tti.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.08 | p-hydroxybenzoate hydroxylase |
| 2bxs.1.A | 26.47 | monomer | HHblits | X-ray | 3.15Å | 0.30 | 0.08 | AMINE OXIDASE [FLAVIN-CONTAINING] A |
| 2yr6.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.35Å | 0.30 | 0.08 | Pro-enzyme of L-phenylalanine oxidase |
| 2yr4.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.70Å | 0.30 | 0.08 | Pro-enzyme of L-phenylalanine oxidase |
| 3rp6.1.A | 21.21 | monomer | HHblits | X-ray | 2.20Å | 0.32 | 0.07 | flavoprotein monooxygenase |
| 3rp8.1.A | 21.21 | monomer | HHblits | X-ray | 1.97Å | 0.32 | 0.07 | flavoprotein monooxygenase |
| 3cge.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.26Å | 0.32 | 0.07 | Pyridine nucleotide-disulfide oxidoreductase, class I |
| 3f3s.1.A | 30.30 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.07 | Lambda-crystallin homolog |
| 3f3s.2.A | 30.30 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.07 | Lambda-crystallin homolog |
| 5wqo.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.78Å | 0.27 | 0.08 | Probable dehydrogenase |
| 5wqp.1.B | 20.00 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.08 | Probable dehydrogenase |
| 1f6m.1.A | 17.14 | hetero-oligomer | HHblits | X-ray | 2.95Å | 0.27 | 0.08 | THIOREDOXIN REDUCTASE |
| 1tdf.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | THIOREDOXIN REDUCTASE |
| 1tde.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.08 | THIOREDOXIN REDUCTASE |
| 1trb.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | THIOREDOXIN REDUCTASE |
| 1cl0.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | THIOREDOXIN REDUCTASE |
| 4u8l.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 4u8o.1.B | 17.14 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 4u8o.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 3utf.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 3utf.1.C | 17.14 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 3uth.1.A | 17.14 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 5vwt.1.B | 17.14 | homo-tetramer | HHblits | X-ray | 2.75Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 1aup.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE |
| 1k89.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.05Å | 0.27 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 1bgv.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 1hrd.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 1.96Å | 0.27 | 0.08 | GLUTAMATE DEHYDROGENASE |
| 5brt.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | 2-hydroxybiphenyl-3-monooxygenase |
| 2raf.1.A | 13.89 | homo-dimer | HHblits | X-ray | 1.60Å | 0.25 | 0.08 | Putative Dinucleotide-Binding Oxidoreductase |
| 3am3.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | Enoyl-ACP reductase |
| 3am5.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.29 | 0.08 | Enoyl-ACP reductase |
| 4bk2.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.47Å | 0.29 | 0.08 | PROBABLE SALICYLATE MONOOXYGENASE |
| 4bjy.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.52Å | 0.29 | 0.08 | PROBABLE SALICYLATE MONOOXYGENASE |
| 4bk1.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.73Å | 0.29 | 0.08 | PROBABLE SALICYLATE MONOOXYGENASE |
| 4bk3.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.78Å | 0.29 | 0.08 | PROBABLE SALICYLATE MONOOXYGENASE |
| 1rz3.1.A | 20.59 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.08 | hypothetical protein RBSTP0775 |
| 2gks.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.31Å | 0.29 | 0.08 | Bifunctional SAT/APS kinase |
| 2gks.1.B | 20.59 | homo-dimer | HHblits | X-ray | 2.31Å | 0.29 | 0.08 | Bifunctional SAT/APS kinase |
| 4rep.1.A | 27.27 | monomer | HHblits | X-ray | 1.97Å | 0.32 | 0.07 | Gamma-carotene desaturase |
| 3rui.1.A | 17.14 | hetero-oligomer | HHblits | X-ray | 1.91Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 3t7e.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 3vh3.1.A | 17.14 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 4gsk.1.A | 17.14 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 4gsl.1.A | 17.14 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 4gsl.1.B | 17.14 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 3vh1.1.A | 17.14 | homo-dimer | HHblits | X-ray | 3.00Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 3vh2.1.A | 17.14 | homo-dimer | HHblits | X-ray | 3.30Å | 0.27 | 0.08 | Ubiquitin-like modifier-activating enzyme ATG7 |
| 3hfs.1.A | 20.00 | monomer | HHblits | X-ray | 3.17Å | 0.27 | 0.08 | Anthocyanidin reductase |
| 2rh8.1.A | 20.00 | monomer | HHblits | X-ray | 2.22Å | 0.27 | 0.08 | Anthocyanidin reductase |
| 1v59.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.08 | Dihydrolipoamide dehydrogenase |
| 1jeh.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1jeh.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 2qa2.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | Polyketide oxygenase CabE |
| 5dbj.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.75Å | 0.27 | 0.08 | FADH2-dependent halogenase PltA |
| 1vqw.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | PROTEIN WITH SIMILARITY TO FLAVIN-CONTAINING MONOOXYGENASES AND TO MAMMALIAN DIMETHYLALANINE MONOOXYGENASES |
| 3aoe.1.D | 17.14 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.27 | 0.08 | Glutamate dehydrogenase |
| 3aoe.1.C | 17.14 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.27 | 0.08 | Glutamate dehydrogenase |
| 3aoe.1.E | 17.14 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.27 | 0.08 | Glutamate dehydrogenase |
| 5jcm.1.A | 17.14 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | Os09g0567300 protein |
| 5jcl.1.A | 17.14 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | Os09g0567300 protein |
| 5m0z.1.A | 8.33 | monomer | HHblits | X-ray | 1.60Å | 0.25 | 0.08 | Cyclohexanone Monooxygenase from Thermocrispum municipale. |
| 1nyt.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.25 | 0.08 | Shikimate 5-dehydrogenase |
| 1nyt.1.C | 13.89 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.25 | 0.08 | Shikimate 5-dehydrogenase |
| 2nvu.1.B | 20.59 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.29 | 0.08 | Maltose binding protein/NEDD8-activating enzyme E1 catalytic subunit chimera |
| 3kyc.1.B | 23.53 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.29 | 0.08 | SUMO-activating enzyme subunit 2 |
| 3upu.1.A | 17.65 | monomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | ATP-dependent DNA helicase dda |
| 3upu.3.A | 17.65 | monomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | ATP-dependent DNA helicase dda |
| 4k6f.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.29 | 0.08 | Putative Acetoacetyl-CoA reductase |
| 4opt.1.A | 23.53 | monomer | HHblits | X-ray | 2.60Å | 0.29 | 0.08 | Conserved Archaeal protein |
| 4opg.1.A | 23.53 | monomer | HHblits | X-ray | 2.07Å | 0.29 | 0.08 | Conserved Archaeal protein |
| 4om8.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.55Å | 0.32 | 0.07 | 3-hydroxybutyryl-coA dehydrogenase |
| 3t61.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.20Å | 0.32 | 0.07 | Gluconokinase |
| 4j34.1.A | 24.24 | monomer | HHblits | X-ray | 2.03Å | 0.32 | 0.07 | Kynurenine 3-monooxygenase |
| 4j34.2.A | 24.24 | monomer | HHblits | X-ray | 2.03Å | 0.32 | 0.07 | Kynurenine 3-monooxygenase |
| 3a4l.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.80Å | 0.32 | 0.07 | L-seryl-tRNA(Sec) kinase |
| 3a4l.1.B | 27.27 | homo-dimer | HHblits | X-ray | 1.80Å | 0.32 | 0.07 | L-seryl-tRNA(Sec) kinase |
| 3am1.1.C | 27.27 | homo-dimer | HHblits | X-ray | 2.40Å | 0.32 | 0.07 | L-seryl-tRNA(Sec) kinase |
| 5o30.1.A | 11.43 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | Putative oxidoreductase |
| 5ijz.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.29Å | 0.27 | 0.08 | NADP-specific glutamate dehydrogenase |
| 5ijz.1.D | 20.00 | homo-hexamer | HHblits | X-ray | 2.29Å | 0.27 | 0.08 | NADP-specific glutamate dehydrogenase |
| 5ijz.1.F | 20.00 | homo-hexamer | HHblits | X-ray | 2.29Å | 0.27 | 0.08 | NADP-specific glutamate dehydrogenase |
| 5ijz.2.F | 20.00 | homo-hexamer | HHblits | X-ray | 2.29Å | 0.27 | 0.08 | NADP-specific glutamate dehydrogenase |
| 2xj9.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.08 | MIPZ |
| 2xj9.1.B | 22.86 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.08 | MIPZ |
| 2ukd.1.A | 20.59 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | URIDYLMONOPHOSPHATE/CYTIDYLMONOPHOSPHATE KINASE |
| 1uke.1.A | 20.59 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | URIDYLMONOPHOSPHATE/CYTIDYLMONOPHOSPHATE KINASE |
| 3umf.1.A | 23.53 | monomer | HHblits | X-ray | 2.05Å | 0.29 | 0.08 | Adenylate kinase |
| 1f48.1.A | 23.53 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | ARSENITE-TRANSLOCATING ATPASE |
| 5lnk.1.Y | 20.59 | hetero-oligomer | HHblits | EM | 3.90Å | 0.29 | 0.08 | Mitochondrial complex I, 42 kDa subunit |
| 3pg5.1.A | 17.65 | monomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Uncharacterized protein |
| 3pg5.2.A | 17.65 | monomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Uncharacterized protein |
| 3pg5.3.A | 17.65 | monomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Uncharacterized protein |
| 3pg5.4.A | 17.65 | monomer | HHblits | X-ray | 3.30Å | 0.29 | 0.08 | Uncharacterized protein |
| 3ihm.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | Styrene monooxygenase A |
| 1qzw.1.B | 27.27 | monomer | HHblits | X-ray | 4.10Å | 0.31 | 0.07 | Signal recognition 54 kDa protein |
| 1qzx.1.A | 27.27 | monomer | HHblits | X-ray | 4.00Å | 0.31 | 0.07 | Signal recognition 54 kDa protein |
| 5l1n.1.A | 20.00 | homo-dimer | HHblits | X-ray | 3.60Å | 0.27 | 0.08 | Coenzyme A disulfide reductase |
| 3kd9.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.75Å | 0.27 | 0.08 | Coenzyme A disulfide reductase |
| 2hdh.1.A | 22.86 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.08 | L-3-HYDROXYACYL COA DEHYDROGENASE |
| 5mzk.2.A | 17.14 | monomer | HHblits | X-ray | 1.82Å | 0.27 | 0.08 | Kynurenine 3-monooxygenase |
| 5fn0.1.A | 17.14 | monomer | HHblits | X-ray | 3.19Å | 0.27 | 0.08 | KYNURENINE 3-MONOOXYGENASE |
| 5mzc.1.A | 17.14 | monomer | HHblits | X-ray | 1.82Å | 0.27 | 0.08 | Kynurenine 3-monooxygenase |
| 5nak.1.B | 17.14 | homo-dimer | HHblits | X-ray | 1.50Å | 0.27 | 0.08 | Kynurenine 3-monooxygenase |
| 5nah.1.A | 17.14 | monomer | HHblits | X-ray | 1.75Å | 0.27 | 0.08 | Kynurenine 3-monooxygenase |
| 4tlz.1.A | 13.89 | homo-tetramer | HHblits | X-ray | 2.41Å | 0.24 | 0.08 | KtzI |
| 1yo6.1.A | 11.11 | homo-dimer | HHblits | X-ray | 2.60Å | 0.24 | 0.08 | Putative Carbonyl Reductase Sniffer |
| 1yo6.3.B | 11.11 | homo-dimer | HHblits | X-ray | 2.60Å | 0.24 | 0.08 | Putative Carbonyl Reductase Sniffer |
| 3orf.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.16Å | 0.29 | 0.08 | Dihydropteridine reductase |
| 3orf.2.A | 17.65 | homo-dimer | HHblits | X-ray | 2.16Å | 0.29 | 0.08 | Dihydropteridine reductase |
| 3cmm.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 3cmm.2.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | Ubiquitin-activating enzyme E1 1 |
| 1zkm.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.95Å | 0.29 | 0.08 | Adenylyltransferase thiF |
| 1zfn.2.B | 23.53 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.08 | Adenylyltransferase thiF |
| 4dim.1.A | 14.71 | monomer | HHblits | X-ray | 2.61Å | 0.29 | 0.08 | Phosphoribosylglycinamide synthetase |
| 1o5w.1.A | 23.53 | homo-dimer | HHblits | X-ray | 3.20Å | 0.29 | 0.08 | Amine oxidase [flavin-containing] A |
| 1o5w.1.B | 23.53 | homo-dimer | HHblits | X-ray | 3.20Å | 0.29 | 0.08 | Amine oxidase [flavin-containing] A |
| 1pxc.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.08 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1pbd.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1cj3.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.08 | PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) |
| 1cc4.1.A | 17.65 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) |
| 1bf3.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1pbf.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.70Å | 0.29 | 0.08 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1bgn.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1bkw.1.A | 17.65 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.08 | PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) |
| 2pey.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.88Å | 0.34 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) |
| 2pey.1.B | 21.88 | homo-dimer | HHblits | X-ray | 1.88Å | 0.34 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) |
| 2ofx.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.90Å | 0.34 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 |
| 1yr6.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.15Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 1yr7.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.08Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 1yr8.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 1yr9.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 1yra.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 1yra.1.B | 21.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 1yrb.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.75Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 2oxr.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.07 | ATP(GTP)binding protein |
| 3ucl.1.A | 27.27 | monomer | HHblits | X-ray | 2.36Å | 0.31 | 0.07 | Cyclohexanone monooxygenase |
| 4rg3.1.A | 27.27 | monomer | HHblits | X-ray | 1.94Å | 0.31 | 0.07 | Cyclohexanone monooxygenase |
| 2dkn.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.07 | 3-alpha-hydroxysteroid dehydrogenase |
| 3h5a.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.07 | MccB protein |
| 3h5a.1.B | 21.21 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.07 | MccB protein |
| 3adk.1.A | 21.21 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | ADENYLATE KINASE |
| 1dxl.1.A | 17.14 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1dxl.1.B | 17.14 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1dxl.2.A | 17.14 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1dxl.2.B | 17.14 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 4gcm.1.A | 17.14 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.08 | Thioredoxin reductase |
| 4jnq.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.08 | Thioredoxin reductase |
| 2yfq.1.A | 14.29 | homo-hexamer | HHblits | X-ray | 2.94Å | 0.26 | 0.08 | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE |
| 5mog.1.A | 17.14 | homo-pentamer | HHblits | X-ray | 2.77Å | 0.26 | 0.08 | Phytoene dehydrogenase, chloroplastic/chromoplastic |
| 2uzz.1.A | 20.59 | monomer | HHblits | X-ray | 3.20Å | 0.29 | 0.08 | N-METHYL-L-TRYPTOPHAN OXIDASE |
| 4e3z.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | Putative oxidoreductase protein |
| 1e2e.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | THYMIDYLATE KINASE |
| 1x31.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.15Å | 0.29 | 0.08 | Sarcosine oxidase alpha subunit |
| 2gah.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.29 | 0.08 | heterotetrameric sarcosine oxidase alpha-subunit |
| 4iv9.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.95Å | 0.29 | 0.08 | Tryptophan 2-monooxygenase |
| 2q99.1.A | 25.00 | monomer | HHblits | X-ray | 1.64Å | 0.33 | 0.07 | Saccharopine dehydrogenase [NAD+, L-lysine-forming |
| 1qo8.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.15Å | 0.31 | 0.07 | FLAVOCYTOCHROME C3 FUMARATE REDUCTASE |
| 5gsn.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.20Å | 0.31 | 0.07 | Flavin-containing monooxygenase |
| 5ipy.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.50Å | 0.31 | 0.07 | Flavin-containing monooxygenase |
| 5iq4.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.50Å | 0.31 | 0.07 | Flavin-containing monooxygenase |
| 2cnw.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.39Å | 0.31 | 0.07 | CELL DIVISION PROTEIN FTSY |
| 2iyl.1.A | 21.21 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | CELL DIVISION PROTEIN FTSY |
| 2cnw.2.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.39Å | 0.31 | 0.07 | CELL DIVISION PROTEIN FTSY |
| 2j7p.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 1.97Å | 0.31 | 0.07 | CELL DIVISION PROTEIN FTSY |
| 1okk.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | CELL DIVISION PROTEIN FTSY |
| 2xkv.1.D | 21.21 | hetero-oligomer | HHblits | EM | 13.50Å | 0.31 | 0.07 | CELL DIVISION PROTEIN FTSY |
| 2q9a.1.A | 21.21 | monomer | HHblits | X-ray | 2.24Å | 0.31 | 0.07 | Cell division protein ftsY |
| 2q9a.2.A | 21.21 | monomer | HHblits | X-ray | 2.24Å | 0.31 | 0.07 | Cell division protein ftsY |
| 5uwy.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.72Å | 0.26 | 0.08 | Thioredoxin reductase |
| 3rnm.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 1zy8.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 1zy8.5.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 1zy8.5.B | 14.29 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 3ucl.1.A | 8.33 | monomer | HHblits | X-ray | 2.36Å | 0.24 | 0.08 | Cyclohexanone monooxygenase |
| 4rg3.1.A | 8.33 | monomer | HHblits | X-ray | 1.94Å | 0.24 | 0.08 | Cyclohexanone monooxygenase |
| 4v03.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | SITE-DETERMINING PROTEIN |
| 2nwq.1.A | 17.65 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | Probable short-chain dehydrogenase |
| 1yr6.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.15Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1yr7.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.08Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1yr8.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1yr9.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.80Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1yra.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.30Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1yra.1.B | 28.13 | homo-dimer | HHblits | X-ray | 2.30Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1yrb.1.A | 28.13 | homo-dimer | HHblits | X-ray | 1.75Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 2oxr.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.07 | ATP(GTP)binding protein |
| 1m7h.4.B | 31.25 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.33 | 0.07 | Adenylylsulfate kinase |
| 1d6j.1.B | 31.25 | homo-dimer | HHblits | X-ray | 2.00Å | 0.33 | 0.07 | ADENOSINE-5'PHOSPHOSULFATE KINASE |
| 1d6j.1.A | 31.25 | homo-dimer | HHblits | X-ray | 2.00Å | 0.33 | 0.07 | ADENOSINE-5'PHOSPHOSULFATE KINASE |
| 1m7h.4.C | 31.25 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.33 | 0.07 | Adenylylsulfate kinase |
| 1m7g.5.A | 31.25 | homo-tetramer | HHblits | X-ray | 1.43Å | 0.33 | 0.07 | Adenylylsulfate kinase |
| 2pk3.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.82Å | 0.31 | 0.07 | GDP-6-deoxy-D-lyxo-4-hexulose reductase |
| 3cm0.1.A | 21.21 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.07 | Adenylate kinase |
| 3kl4.1.A | 27.27 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.31 | 0.07 | Signal recognition 54 kDa protein |
| 4qec.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.08 | ElxO |
| 4qed.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.08 | ElxO |
| 3r75.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | Anthranilate/para-aminobenzoate synthases component I |
| 3r75.1.B | 14.29 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | Anthranilate/para-aminobenzoate synthases component I |
| 3r74.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.90Å | 0.26 | 0.08 | Anthranilate/para-aminobenzoate synthases component I |
| 3r74.1.B | 14.29 | homo-dimer | HHblits | X-ray | 2.90Å | 0.26 | 0.08 | Anthranilate/para-aminobenzoate synthases component I |
| 4ivo.1.A | 14.29 | monomer | HHblits | X-ray | 2.60Å | 0.26 | 0.08 | Protoporphyrinogen oxidase |
| 4ivm.1.A | 14.29 | monomer | HHblits | X-ray | 2.77Å | 0.26 | 0.08 | Protoporphyrinogen oxidase |
| 2bma.1.A | 14.29 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.26 | 0.08 | GLUTAMATE DEHYDROGENASE (NADP+) |
| 1dts.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.65Å | 0.28 | 0.08 | DETHIOBIOTIN SYNTHETASE |
| 1dah.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.64Å | 0.28 | 0.08 | DETHIOBIOTIN SYNTHETASE |
| 1byi.1.A | 20.59 | homo-dimer | HHblits | X-ray | 0.97Å | 0.28 | 0.08 | DETHIOBIOTIN SYNTHASE |
| 1cj4.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) |
| 2if2.1.A | 31.25 | monomer | HHblits | X-ray | 3.00Å | 0.33 | 0.07 | Dephospho-CoA kinase |
| 2if2.2.A | 31.25 | monomer | HHblits | X-ray | 3.00Å | 0.33 | 0.07 | Dephospho-CoA kinase |
| 2if2.3.A | 31.25 | monomer | HHblits | X-ray | 3.00Å | 0.33 | 0.07 | Dephospho-CoA kinase |
| 2ax4.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.50Å | 0.31 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 |
| 3ndp.1.A | 21.21 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | Adenylate kinase isoenzyme 4 |
| 5eow.1.A | 21.21 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | 6-hydroxynicotinate 3-monooxygenase |
| 1p2e.1.A | 27.27 | monomer | HHblits | X-ray | 2.20Å | 0.31 | 0.07 | flavocytochrome c3 |
| 2b7r.1.A | 27.27 | monomer | HHblits | X-ray | 1.70Å | 0.31 | 0.07 | Fumarate reductase flavoprotein subunit |
| 1p2h.1.A | 27.27 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | flavocytochrome c3 |
| 1zx9.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | Mercuric reductase |
| 4k7z.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.50Å | 0.31 | 0.07 | Mercuric reductase |
| 4h4z.1.A | 14.29 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.08 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 3fkq.1.A | 5.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | NtrC-like two-domain protein |
| 5j7x.1.A | 8.33 | monomer | HHblits | X-ray | 1.90Å | 0.24 | 0.08 | Dimethylaniline monooxygenase, putative |
| 3lw7.1.A | 35.48 | homo-dimer | HHblits | X-ray | 2.30Å | 0.35 | 0.07 | Adenylate kinase related protein (AdkA-like) |
| 3lw7.1.B | 35.48 | homo-dimer | HHblits | X-ray | 2.30Å | 0.35 | 0.07 | Adenylate kinase related protein (AdkA-like) |
| 2f6r.1.A | 38.71 | monomer | HHblits | X-ray | 1.70Å | 0.35 | 0.07 | Bifunctional coenzyme A synthase |
| 2duw.1.A | 17.65 | monomer | HHblits | NMR | NA | 0.28 | 0.08 | putative CoA-binding protein |
| 4fgw.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.45Å | 0.28 | 0.08 | Glycerol-3-phosphate dehydrogenase [NAD(+)] 1 |
| 1vl0.1.A | 14.71 | homo-trimer | HHblits | X-ray | 2.05Å | 0.28 | 0.08 | DTDP-4-dehydrorhamnose reductase, rfbD ortholog |
| 2ylr.1.A | 23.53 | monomer | HHblits | X-ray | 2.26Å | 0.28 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 1w4x.1.A | 23.53 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 2ym2.1.A | 23.53 | monomer | HHblits | X-ray | 2.70Å | 0.28 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 2ylz.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 4d03.1.A | 23.53 | monomer | HHblits | X-ray | 1.81Å | 0.28 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 2ylx.1.A | 23.53 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.08 | PHENYLACETONE MONOOXYGENASE |
| 1cke.1.A | 17.65 | monomer | HHblits | X-ray | 1.75Å | 0.28 | 0.08 | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE) |
| 1rp0.1.A | 17.65 | homo-octamer | HHblits | X-ray | 1.60Å | 0.28 | 0.08 | Thiazole biosynthetic enzyme |
| 3gwf.1.A | 8.33 | monomer | HHblits | X-ray | 2.20Å | 0.24 | 0.08 | Cyclohexanone monooxygenase |
| 3gwd.1.A | 8.33 | monomer | HHblits | X-ray | 2.30Å | 0.24 | 0.08 | Cyclohexanone monooxygenase |
| 5mq6.1.A | 8.57 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.08 | Pyridine nucleotide-disulfide oxidoreductase-like protein |
| 4id9.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.60Å | 0.33 | 0.07 | SHORT-CHAIN DEHYDROGENASE/REDUCTASE |
| 3cwq.1.A | 37.50 | homo-dimer | HHblits | X-ray | 2.47Å | 0.33 | 0.07 | ParA family chromosome partitioning protein |
| 5mq6.1.A | 24.24 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.07 | Pyridine nucleotide-disulfide oxidoreductase-like protein |
| 4r1n.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.07 | 3-hydroxybutyryl-CoA dehydrogenase |
| 4r1n.2.B | 21.21 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.07 | 3-hydroxybutyryl-CoA dehydrogenase |
| 3trf.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.60Å | 0.30 | 0.07 | Shikimate kinase |
| 3jaj.45.A | 27.27 | monomer | HHblits | EM | NA | 0.30 | 0.07 | SRP54 |
| 3jan.45.A | 27.27 | monomer | HHblits | EM | NA | 0.30 | 0.07 | SRP54 |
| 3mle.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.07 | Dethiobiotin synthetase |
| 3lb8.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | Putidaredoxin reductase |
| 3lb8.2.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | Putidaredoxin reductase |
| 1q1r.1.A | 20.59 | monomer | HHblits | X-ray | 1.91Å | 0.28 | 0.08 | Putidaredoxin reductase |
| 1q1w.2.A | 20.59 | monomer | HHblits | X-ray | 2.60Å | 0.28 | 0.08 | Putidaredoxin reductase |
| 3u62.1.A | 14.71 | monomer | HHblits | X-ray | 1.45Å | 0.28 | 0.08 | Shikimate dehydrogenase |
| 1wpq.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic |
| 4v02.1.A | 17.65 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.28 | 0.08 | SITE-DETERMINING PROTEIN |
| 4jdr.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.50Å | 0.28 | 0.08 | Dihydrolipoyl dehydrogenase |
| 1gt8.1.A | 14.71 | homo-dimer | HHblits | X-ray | 3.30Å | 0.28 | 0.08 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1gth.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.25Å | 0.28 | 0.08 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1h7w.2.A | 14.71 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1h7x.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.01Å | 0.28 | 0.08 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 4u7w.1.A | 13.89 | homo-dimer | HHblits | X-ray | 1.90Å | 0.23 | 0.08 | MxaA |
| 3gzn.1.A | 11.43 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.26 | 0.08 | NEDD8-activating enzyme E1 regulatory subunit |
| 2nvu.1.A | 11.43 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.26 | 0.08 | NEDD8-activating enzyme E1 regulatory subunit |
| 3dbh.1.A | 11.43 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.26 | 0.08 | NEDD8-activating enzyme E1 regulatory subunit |
| 1tt5.1.A | 11.43 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.26 | 0.08 | amyloid protein-binding protein 1 |
| 1tt5.2.A | 11.43 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.26 | 0.08 | amyloid protein-binding protein 1 |
| 1r4m.1.A | 11.43 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.26 | 0.08 | amyloid beta precursor protein-binding protein 1 |
| 1yov.1.A | 11.43 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.26 | 0.08 | Amyloid protein-binding protein 1 |
| 1yov.2.A | 11.43 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.26 | 0.08 | Amyloid protein-binding protein 1 |
| 4qqr.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.08 | 3,5-epimerase/4-reductase |
| 3dfu.1.A | 17.14 | homo-dimer | HHblits | X-ray | 2.07Å | 0.26 | 0.08 | Uncharacterized Protein from 6-Phosphogluconate Dehydrogenase-Like Family |
| 3dfu.1.B | 17.14 | homo-dimer | HHblits | X-ray | 2.07Å | 0.26 | 0.08 | Uncharacterized Protein from 6-Phosphogluconate Dehydrogenase-Like Family |
| 3vzq.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.30 | 0.07 | Acetoacetyl-CoA reductase |
| 3vzp.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 1.79Å | 0.30 | 0.07 | Acetoacetyl-CoA reductase |
| 2pey.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.88Å | 0.30 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) |
| 2pey.1.B | 24.24 | homo-dimer | HHblits | X-ray | 1.88Å | 0.30 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) |
| 2ofx.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 |
| 2i0z.1.A | 27.27 | monomer | HHblits | X-ray | 1.84Å | 0.30 | 0.07 | NAD(FAD)-utilizing dehydrogenases |
| 2z5y.1.A | 27.27 | monomer | HHblits | X-ray | 2.17Å | 0.30 | 0.07 | Amine oxidase [flavin-containing] A |
| 2z5x.1.A | 27.27 | monomer | HHblits | X-ray | 2.20Å | 0.30 | 0.07 | Amine oxidase [flavin-containing] A |
| 3k30.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.07 | Histamine dehydrogenase |
| 1jjv.1.A | 21.88 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.07 | DEPHOSPHO-COA KINASE |
| 5wqn.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | Probable dehydrogenase |
| 5wqn.1.B | 20.59 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | Probable dehydrogenase |
| 5wqn.2.B | 20.59 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.08 | Probable dehydrogenase |
| 3tf5.1.A | 17.65 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 4edf.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.08Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3itk.1.A | 17.65 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 3itk.1.F | 17.65 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 5tjh.1.A | 17.65 | homo-hexamer | HHblits | X-ray | 2.05Å | 0.28 | 0.08 | UDP-glucose 6-dehydrogenase |
| 4e12.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.93Å | 0.28 | 0.08 | Diketoreductase |
| 4dsh.1.A | 17.65 | monomer | HHblits | X-ray | 2.25Å | 0.28 | 0.08 | UDP-galactopyranose mutase |
| 1zmd.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.08Å | 0.28 | 0.08 | Dihydrolipoyl dehydrogenase |
| 2bry.1.A | 23.53 | monomer | HHblits | X-ray | 1.45Å | 0.28 | 0.08 | NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS |
| 2c4c.2.A | 23.53 | monomer | HHblits | X-ray | 2.90Å | 0.28 | 0.08 | NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS |
| 2c4c.1.A | 23.53 | monomer | HHblits | X-ray | 2.90Å | 0.28 | 0.08 | NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS |
| 3k6j.1.A | 11.43 | monomer | HHblits | X-ray | 2.20Å | 0.25 | 0.08 | Protein F01G10.3, confirmed by transcript evidence |
| 3sbo.1.D | 17.14 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.25 | 0.08 | NADP-specific glutamate dehydrogenase |
| 4bht.1.E | 17.14 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.25 | 0.08 | NADP-SPECIFIC GLUTAMATE DEHYDROGENASE |
| 4bht.1.D | 17.14 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.25 | 0.08 | NADP-SPECIFIC GLUTAMATE DEHYDROGENASE |
| 4bht.1.C | 17.14 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.25 | 0.08 | NADP-SPECIFIC GLUTAMATE DEHYDROGENASE |
| 3sbo.1.F | 17.14 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.25 | 0.08 | NADP-specific glutamate dehydrogenase |
| 4bht.1.F | 17.14 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.25 | 0.08 | NADP-SPECIFIC GLUTAMATE DEHYDROGENASE |
| 3sbo.1.E | 17.14 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.25 | 0.08 | NADP-specific glutamate dehydrogenase |
| 3sbo.1.C | 17.14 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.25 | 0.08 | NADP-specific glutamate dehydrogenase |
| 4fhn.3.C | 17.14 | homo-hexamer | HHblits | X-ray | 6.99Å | 0.25 | 0.08 | Glutamate dehydrogenase |
| 4fcc.1.E | 17.14 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.25 | 0.08 | Glutamate dehydrogenase |
| 4fcc.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.25 | 0.08 | Glutamate dehydrogenase |
| 4fcc.1.B | 17.14 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.25 | 0.08 | Glutamate dehydrogenase |
| 2yfh.1.A | 17.14 | homo-hexamer | HHblits | X-ray | 2.69Å | 0.25 | 0.08 | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, GLUTAMATE DEHYDROGENASE |
| 2yfh.1.C | 17.14 | homo-hexamer | HHblits | X-ray | 2.69Å | 0.25 | 0.08 | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, GLUTAMATE DEHYDROGENASE |
| 2yfh.1.D | 17.14 | homo-hexamer | HHblits | X-ray | 2.69Å | 0.25 | 0.08 | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, GLUTAMATE DEHYDROGENASE |
| 2yfh.1.E | 17.14 | homo-hexamer | HHblits | X-ray | 2.69Å | 0.25 | 0.08 | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, GLUTAMATE DEHYDROGENASE |
| 2yfh.1.F | 17.14 | homo-hexamer | HHblits | X-ray | 2.69Å | 0.25 | 0.08 | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, GLUTAMATE DEHYDROGENASE |
| 4c0h.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.25 | 0.08 | MRNA CLEAVAGE AND POLYADENYLATION FACTOR CLP1 |
| 1zmd.1.A | 11.43 | homo-dimer | HHblits | X-ray | 2.08Å | 0.25 | 0.08 | Dihydrolipoyl dehydrogenase |
| 1ko7.1.A | 27.27 | homo-trimer | HHblits | X-ray | 1.95Å | 0.30 | 0.07 | Hpr kinase/phosphatase |
| 1ko7.2.A | 27.27 | homo-trimer | HHblits | X-ray | 1.95Å | 0.30 | 0.07 | Hpr kinase/phosphatase |
| 4zcc.1.B | 24.24 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.07 | Renalase |
| 4zcc.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.07 | Renalase |
| 4zcd.3.A | 24.24 | monomer | HHblits | X-ray | 1.66Å | 0.30 | 0.07 | Renalase |
| 4zcd.2.A | 24.24 | monomer | HHblits | X-ray | 1.66Å | 0.30 | 0.07 | Renalase |
| 1x31.1.B | 27.27 | hetero-oligomer | HHblits | X-ray | 2.15Å | 0.30 | 0.07 | Sarcosine oxidase beta subunit |
| 3ad9.1.B | 27.27 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.30 | 0.07 | SARCOSINE OXIDASE BETA SUBUNIT |
| 3cty.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.35Å | 0.30 | 0.07 | Thioredoxin reductase |
| 2ax4.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.50Å | 0.32 | 0.07 | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 |
| 3cgb.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.32 | 0.07 | Pyridine nucleotide-disulfide oxidoreductase, class I |
| 2pv7.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | T-protein [Includes: Chorismate mutase (EC 5.4.99.5) (CM) and Prephenate dehydrogenase (EC 1.3.1.12) (PDH)] |
| 2nq8.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | Enoyl-acyl carrier reductase |
| 2b76.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.27 | 0.08 | Fumarate reductase flavoprotein subunit |
| 2b76.2.A | 14.71 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.27 | 0.08 | Fumarate reductase flavoprotein subunit |
| 2ydx.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.08 | METHIONINE ADENOSYLTRANSFERASE 2 SUBUNIT BETA |
| 2ydy.1.A | 11.76 | monomer | HHblits | X-ray | 2.25Å | 0.27 | 0.08 | METHIONINE ADENOSYLTRANSFERASE 2 SUBUNIT BETA |
| 1f48.1.A | 14.71 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | ARSENITE-TRANSLOCATING ATPASE |
| 3ay6.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.08 | Glucose 1-dehydrogenase 4 |
| 3ay7.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | Glucose 1-dehydrogenase 4 |
| 4z2u.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | 2-hydroxybiphenyl-3-monooxygenase |
| 4z2r.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | 2-hydroxybiphenyl-3-monooxygenase |
| 4cy6.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 2.76Å | 0.27 | 0.08 | 2-HYDROXYBIPHENYL-3-MONOOXYGENASE |
| 4cy8.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 2.03Å | 0.27 | 0.08 | 2-HYDROXYBIPHENYL 3-MONOOXYGENASE |
| 5uao.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.88Å | 0.27 | 0.08 | Tryptophane-5-halogenase |
| 3i4f.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.39Å | 0.25 | 0.08 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5bt9.1.A | 17.14 | monomer | HHblits | X-ray | 1.50Å | 0.25 | 0.08 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 4w9n.1.A | 11.43 | homo-dimer | HHblits | X-ray | 1.84Å | 0.25 | 0.08 | Enoyl-[acyl-carrier-protein] reductase |
| 4w82.1.A | 11.43 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.08 | Fatty acid synthase |
| 4lu6.1.A | 14.29 | homo-dimer | HHblits | X-ray | 3.05Å | 0.25 | 0.08 | Flavin-dependent tryptophan halogenase RebH |
| 2iys.1.A | 29.03 | monomer | HHblits | X-ray | 1.40Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 2iyv.1.A | 29.03 | monomer | HHblits | X-ray | 1.35Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 2iyx.1.A | 29.03 | monomer | HHblits | X-ray | 1.49Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 2iyq.1.A | 29.03 | monomer | HHblits | X-ray | 1.80Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 2iyr.1.A | 29.03 | monomer | HHblits | X-ray | 1.98Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 2iyr.2.A | 29.03 | monomer | HHblits | X-ray | 1.98Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 1we2.1.A | 29.03 | monomer | HHblits | X-ray | 2.30Å | 0.35 | 0.07 | Shikimate kinase |
| 1zyu.1.A | 29.03 | monomer | HHblits | X-ray | 2.90Å | 0.35 | 0.07 | Shikimate kinase |
| 3baf.1.A | 29.03 | monomer | HHblits | X-ray | 2.25Å | 0.35 | 0.07 | Shikimate kinase |
| 2g1j.1.A | 29.03 | monomer | HHblits | X-ray | 2.00Å | 0.35 | 0.07 | Shikimate kinase |
| 2g1j.2.A | 29.03 | monomer | HHblits | X-ray | 2.00Å | 0.35 | 0.07 | Shikimate kinase |
| 2dfn.1.A | 29.03 | monomer | HHblits | X-ray | 1.93Å | 0.35 | 0.07 | Shikimate kinase |
| 2dft.1.A | 29.03 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.35 | 0.07 | Shikimate kinase |
| 2dft.1.B | 29.03 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.35 | 0.07 | Shikimate kinase |
| 2dft.1.C | 29.03 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.35 | 0.07 | Shikimate kinase |
| 4bqs.2.A | 29.03 | monomer | HHblits | X-ray | 2.15Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 4bqs.3.A | 29.03 | monomer | HHblits | X-ray | 2.15Å | 0.35 | 0.07 | SHIKIMATE KINASE |
| 2raa.1.A | 30.30 | monomer | HHblits | X-ray | 2.12Å | 0.30 | 0.07 | Pyruvate synthase subunit porC |
| 1v35.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.07 | enoyl-ACP reductase |
| 3kkj.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.50Å | 0.30 | 0.07 | Amine oxidase, flavin-containing |
| 4iqg.1.A | 24.24 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.30 | 0.07 | Short-chain dehydrogenase/reductase SDR |
| 1zcj.1.A | 15.15 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.07 | Peroxisomal bifunctional enzyme |
| 1rz3.1.A | 28.13 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.07 | hypothetical protein RBSTP0775 |
| 3gg2.1.A | 31.25 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.32 | 0.07 | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family |
| 2wtb.1.A | 17.65 | monomer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | FATTY ACID MULTIFUNCTIONAL PROTEIN (ATMFP2) |
| 4wok.1.A | 26.47 | monomer | HHblits | X-ray | 1.85Å | 0.27 | 0.08 | UDP-glucose 4-epimerase |
| 3had.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) |
| 1lsj.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | 3-HYDROXYACYL-COA DEHYDROGENASE |
| 1m76.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.08 | 3-HYDROXYACYL-COA DEHYDROGENASE |
| 1il0.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.08 | 3-hydroxyacyl-CoA dehydrogenase |
| 1f14.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | L-3-HYDROXYACYL-COA DEHYDROGENASE |
| 1f0y.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | L-3-HYDROXYACYL-COA DEHYDROGENASE |
| 3rqs.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial |
| 1m75.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | 3-HYDROXYACYL-COA DEHYDROGENASE |
| 3hdh.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.08 | PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) |
| 3hdh.1.B | 23.53 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.08 | PROTEIN (L-3-HYDROXYACYL COA DEHYDROGENASE) |
| 1lso.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.60Å | 0.27 | 0.08 | 3-HYDROXYACYL-COA DEHYDROGENASE |
| 4p4l.1.A | 14.71 | monomer | HHblits | X-ray | 2.01Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase AroE (5-dehydroshikimate reductase) |
| 4p4g.2.A | 14.71 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase AroE (5-dehydroshikimate reductase) |
| 4p4g.1.A | 14.71 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.08 | Shikimate 5-dehydrogenase AroE (5-dehydroshikimate reductase) |
| 4icy.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | Polyketide oxygenase PgaE |
| 2qa1.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.08 | Polyketide oxygenase PgaE |
| 2gv8.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.08 | monooxygenase |
| 6aon.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.72Å | 0.27 | 0.08 | Dihydrolipoyl dehydrogenase |
| 1wam.1.A | 30.30 | monomer | HHblits | X-ray | 2.35Å | 0.29 | 0.07 | UDP-GALACTOPYRANOSE MUTASE |
| 2bi7.1.A | 30.30 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | UDP-GALACTOPYRANOSE MUTASE |
| 3gf4.1.A | 30.30 | homo-dimer | HHblits | X-ray | 2.45Å | 0.29 | 0.07 | UDP-galactopyranose mutase |
| 3gf4.1.B | 30.30 | homo-dimer | HHblits | X-ray | 2.45Å | 0.29 | 0.07 | UDP-galactopyranose mutase |
| 3int.1.B | 30.30 | homo-dimer | HHblits | X-ray | 2.51Å | 0.29 | 0.07 | Probable UDP-galactopyranose mutase |
| 2xve.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.99Å | 0.29 | 0.07 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xvh.1.B | 18.18 | homo-dimer | HHblits | X-ray | 2.54Å | 0.29 | 0.07 | FLAVIN-CONTAINING MONOOXYGENASE |
| 2xvj.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.48Å | 0.29 | 0.07 | FLAVIN-CONTAINING MONOOXYGENASE |
| 1bgj.1.A | 18.18 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1cc6.1.A | 18.18 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.07 | PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) |
| 3g5q.1.A | 21.21 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO |
| 3g5s.1.A | 21.21 | monomer | HHblits | X-ray | 1.05Å | 0.29 | 0.07 | Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO |
| 3g5r.1.A | 21.21 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO |
| 5l1n.1.A | 21.21 | homo-dimer | HHblits | X-ray | 3.60Å | 0.29 | 0.07 | Coenzyme A disulfide reductase |
| 4fx9.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.70Å | 0.29 | 0.07 | Coenzyme A disulfide reductase |
| 3kd9.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.07 | Coenzyme A disulfide reductase |
| 3zn2.1.A | 31.25 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.32 | 0.07 | HALOHYDRIN DEHALOGENASE |
| 4ixt.1.A | 31.25 | homo-tetramer | HHblits | X-ray | 2.49Å | 0.32 | 0.07 | Halohydrin dehalogenase |
| 1zo8.1.A | 31.25 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.32 | 0.07 | halohydrin dehalogenase |
| 1pj5.1.A | 20.59 | homo-tetramer | HHblits | X-ray | 1.61Å | 0.27 | 0.08 | N,N-dimethylglycine oxidase |
| 5eqd.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.83Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 5eqd.1.B | 23.53 | homo-dimer | HHblits | X-ray | 1.83Å | 0.27 | 0.08 | UDP-galactopyranose mutase |
| 2ive.1.A | 20.59 | monomer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | PROTOPORPHYRINOGEN OXIDASE |
| 5m0z.1.A | 17.65 | monomer | HHblits | X-ray | 1.60Å | 0.27 | 0.08 | Cyclohexanone Monooxygenase from Thermocrispum municipale. |
| 2eq8.1.A | 17.65 | hetero-oligomer | HHblits | X-ray | 1.94Å | 0.27 | 0.08 | Pyruvate dehydrogenase complex, dihydrolipoamide dehydrogenase E3 component |
| 2hqm.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.08 | Glutathione reductase |
| 4oyh.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.2.A | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.2.B | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.3.A | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.3.B | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.2.A | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.2.B | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.3.A | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.3.B | 17.65 | homo-dimer | HHblits | X-ray | 2.41Å | 0.27 | 0.08 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 3tqf.1.A | 24.24 | homo-trimer | HHblits | X-ray | 2.80Å | 0.29 | 0.07 | Hpr(Ser) kinase |
| 3dbr.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 3.05Å | 0.29 | 0.07 | NEDD8-activating enzyme E1 catalytic subunit |
| 3dbh.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.85Å | 0.29 | 0.07 | NEDD8-activating enzyme E1 catalytic subunit |
| 1tt5.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.29 | 0.07 | ubiquitin-activating enzyme E1C isoform 1 |
| 1tt5.2.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.29 | 0.07 | ubiquitin-activating enzyme E1C isoform 1 |
| 3nlc.1.A | 24.24 | monomer | HHblits | X-ray | 2.15Å | 0.29 | 0.07 | Uncharacterized protein VP0956 |
| 3i3l.1.A | 21.21 | monomer | HHblits | X-ray | 2.20Å | 0.29 | 0.07 | Alkylhalidase CmlS |
| 5x1y.1.A | 24.24 | homo-dimer | HHblits | X-ray | 3.48Å | 0.29 | 0.07 | Mercuric reductase |
| 5fjm.1.A | 21.21 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | L-AMINO ACID DEAMINASE |
| 1q3t.1.A | 18.18 | monomer | HHblits | NMR | NA | 0.29 | 0.07 | Cytidylate kinase |
| 2bwj.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.07 | ADENYLATE KINASE 5 |
| 2bwj.1.B | 25.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.07 | ADENYLATE KINASE 5 |
| 2bwj.2.B | 25.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.07 | ADENYLATE KINASE 5 |
| 4wpg.1.A | 21.88 | monomer | HHblits | X-ray | 1.10Å | 0.32 | 0.07 | dTDP-4-dehydrorhamnose reductase |
| 3uwo.1.A | 28.13 | monomer | HHblits | X-ray | 1.70Å | 0.32 | 0.07 | Thymidylate kinase |
| 2jat.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.60Å | 0.32 | 0.07 | DEOXYGUANOSINE KINASE |
| 3uxm.1.A | 28.13 | monomer | HHblits | X-ray | 1.95Å | 0.32 | 0.07 | Thymidylate kinase |
| 3uxm.2.A | 28.13 | monomer | HHblits | X-ray | 1.95Å | 0.32 | 0.07 | Thymidylate kinase |
| 2ol4.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 2.26Å | 0.27 | 0.08 | Enoyl-acyl carrier reductase |
| 4k22.1.A | 23.53 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | Protein VisC |
| 5mh4.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.14Å | 0.27 | 0.08 | Thioredoxin reductase |
| 5mjk.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.08 | Thioredoxin reductase |
| 3vrd.1.B | 14.71 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.27 | 0.08 | Flavocytochrome c flavin subunit |
| 5fs6.1.A | 11.76 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.08 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs7.1.A | 11.76 | monomer | HHblits | X-ray | 1.85Å | 0.27 | 0.08 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs9.1.A | 11.76 | monomer | HHblits | X-ray | 1.75Å | 0.27 | 0.08 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5jci.1.A | 17.65 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.08 | Os09g0567300 protein |
| 1zw1.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.29 | 0.07 | enoyl-acyl carrier reductase |
| 1zsn.1.B | 18.18 | homo-tetramer | HHblits | X-ray | 2.99Å | 0.29 | 0.07 | enoyl-acyl carrier reductase |
| 3lov.1.A | 21.21 | monomer | HHblits | X-ray | 2.06Å | 0.29 | 0.07 | Protoporphyrinogen oxidase |
| 1k0i.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1dob.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1ykj.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | P-hydroxybenzoate hydroxylase |
| 1ykj.1.B | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | P-hydroxybenzoate hydroxylase |
| 1doe.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1cj2.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.07 | PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) |
| 1pxa.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 2qg6.1.A | 25.00 | monomer | HHblits | X-ray | 1.50Å | 0.31 | 0.07 | Nicotinamide riboside kinase 1 |
| 2grj.1.A | 25.00 | monomer | HHblits | X-ray | 2.60Å | 0.31 | 0.07 | Dephospho-CoA kinase |
| 4i1v.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.60Å | 0.31 | 0.07 | Dephospho-CoA kinase |
| 4i1u.1.B | 21.88 | homo-dimer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | Dephospho-CoA kinase |
| 4i1u.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | Dephospho-CoA kinase |
| 3l8k.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | Dihydrolipoyl dehydrogenase |
| 4nwz.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.08 | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| 5kmp.1.A | 14.71 | homo-dimer | HHblits | X-ray | 3.20Å | 0.27 | 0.08 | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| 5kmq.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.70Å | 0.27 | 0.08 | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| 4z44.1.A | 20.59 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.08 | Flavin-dependent tryptophan halogenase PrnA |
| 5gmo.1.A | 14.71 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.08 | Protein induced by osmotic stress |
| 3ef6.1.A | 18.18 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Toluene 1,2-dioxygenase system ferredoxin--NAD(+) reductase |
| 4emi.1.A | 18.18 | monomer | HHblits | X-ray | 1.81Å | 0.29 | 0.07 | TodA |
| 4emj.1.A | 18.18 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.29 | 0.07 | TodA |
| 3sxi.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.18Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3t2y.1.A | 21.21 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3szf.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3sx6.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3kpk.1.A | 21.21 | monomer | HHblits | X-ray | 2.05Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3szw.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 2wdq.1.A | 21.21 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.29 | 0.07 | SUCCINATE DEHYDROGENASE FLAVOPROTEIN SUBUNIT |
| 2acz.1.A | 21.21 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.29 | 0.07 | Succinate dehydrogenase flavoprotein subunit |
| 3syi.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3sz0.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.15Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3t2z.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | Sulfide-quinone reductase, putative |
| 3m0o.1.A | 24.24 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | Monomeric sarcosine oxidase |
| 2a89.1.A | 24.24 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.07 | Monomeric sarcosine oxidase |
| 4dgk.1.A | 21.21 | monomer | HHblits | X-ray | 2.35Å | 0.29 | 0.07 | Phytoene dehydrogenase |
| 3m13.1.A | 24.24 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Monomeric sarcosine oxidase |
| 1l9c.1.A | 24.24 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | Monomeric Sarcosine Oxidase |
| 3bhf.1.A | 24.24 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Monomeric sarcosine oxidase |
| 1zov.1.A | 24.24 | monomer | HHblits | X-ray | 1.86Å | 0.29 | 0.07 | Monomeric sarcosine oxidase |
| 4b63.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | L-ORNITHINE N5 MONOOXYGENASE |
| 3aw8.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.07 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 3jsk.1.A | 24.24 | homo-octamer | HHblits | X-ray | 2.70Å | 0.29 | 0.07 | CyPBP37 protein |
| 3r20.1.A | 21.21 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | Cytidylate kinase |
| 2ofw.1.B | 22.58 | homo-dimer | HHblits | X-ray | 2.05Å | 0.34 | 0.07 | APS kinase domain of the PAPS synthetase 1 |
| 2ofw.1.A | 22.58 | homo-dimer | HHblits | X-ray | 2.05Å | 0.34 | 0.07 | APS kinase domain of the PAPS synthetase 1 |
| 4oyh.1.A | 29.03 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.2.A | 29.03 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.2.B | 29.03 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.3.A | 29.03 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4oyh.3.B | 29.03 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1rj9.1.A | 21.88 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | Signal Recognition Protein |
| 4qtz.1.A | 14.71 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.08 | Dihydroflavonol-4-reductase |
| 1fcd.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 2.53Å | 0.26 | 0.08 | FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT) |
| 1fcd.2.A | 14.71 | hetero-oligomer | HHblits | X-ray | 2.53Å | 0.26 | 0.08 | FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT) |
| 3l6e.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 3l6e.1.B | 24.24 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 3asv.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.29 | 0.07 | Short-chain dehydrogenase/reductase SDR |
| 4z9f.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.29 | 0.07 | Halohydrin epoxidase A |
| 4ap1.1.A | 21.21 | monomer | HHblits | X-ray | 2.95Å | 0.29 | 0.07 | STEROID MONOOXYGENASE |
| 4ap3.1.A | 21.21 | monomer | HHblits | X-ray | 2.39Å | 0.29 | 0.07 | STEROID MONOOXYGENASE |
| 4aos.1.A | 21.21 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | STEROID MONOOXYGENASE |
| 1m66.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | Glycerol-3-phosphate dehydrogenase |
| 1n1e.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | glycerol-3-phosphate dehydrogenase |
| 1e9c.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | THYMIDYLATE KINASE |
| 1e98.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | THYMIDYLATE KINASE |
| 2xx3.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | THYMIDYLATE KINASE |
| 5i39.1.A | 27.27 | monomer | HHblits | X-ray | 1.20Å | 0.29 | 0.07 | L-amino acid deaminase |
| 5hxw.1.A | 27.27 | monomer | HHblits | X-ray | 2.63Å | 0.29 | 0.07 | L-amino acid deaminase |
| 4ol9.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.85Å | 0.29 | 0.07 | Putative 2-dehydropantoate 2-reductase |
| 2pbr.1.B | 38.71 | homo-dimer | HHblits | X-ray | 1.96Å | 0.34 | 0.07 | Thymidylate kinase |
| 2pbr.1.A | 38.71 | homo-dimer | HHblits | X-ray | 1.96Å | 0.34 | 0.07 | Thymidylate kinase |
| 4s35.1.B | 38.71 | homo-dimer | HHblits | X-ray | 1.55Å | 0.34 | 0.07 | Thymidylate kinase |
| 5xb2.1.B | 38.71 | homo-dimer | HHblits | X-ray | 2.16Å | 0.34 | 0.07 | Thymidylate kinase |
| 5xbh.1.A | 38.71 | homo-dimer | HHblits | X-ray | 2.23Å | 0.34 | 0.07 | Thymidylate kinase |
| 2ofw.1.B | 28.13 | homo-dimer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | APS kinase domain of the PAPS synthetase 1 |
| 2ofw.1.A | 28.13 | homo-dimer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | APS kinase domain of the PAPS synthetase 1 |
| 5d1f.1.A | 21.88 | homo-dimer | HHblits | X-ray | 3.40Å | 0.31 | 0.07 | Pyruvate, phosphate dikinase regulatory protein, chloroplastic |
| 1mo9.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.65Å | 0.26 | 0.08 | orf3 |
| 1mok.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.08 | orf3 |
| 1sny.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.75Å | 0.26 | 0.08 | sniffer CG10964-PA |
| 2yvg.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.60Å | 0.26 | 0.08 | Ferredoxin reductase |
| 2yvj.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.26 | 0.08 | Ferredoxin reductase |
| 2yvj.1.C | 14.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.26 | 0.08 | Ferredoxin reductase |
| 1d7y.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | FERREDOXIN REDUCTASE |
| 2gr3.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.08 | ferredoxin reductase |
| 4h4p.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.08 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4h4x.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.08 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4h4t.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.08 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4h4r.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.40Å | 0.26 | 0.08 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4h4v.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.40Å | 0.26 | 0.08 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 1m6v.1.A | 17.65 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | carbamoyl phosphate synthetase large chain |
| 2h11.1.A | 14.71 | monomer | HHblits | X-ray | 1.89Å | 0.26 | 0.08 | Thiopurine S-methyltransferase |
| 1x0v.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic |
| 1x0x.1.A | 21.21 | monomer | HHblits | X-ray | 2.75Å | 0.28 | 0.07 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic |
| 4o6v.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.07 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 2qq0.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.28 | 0.07 | Thymidine kinase |
| 2qpo.1.B | 15.15 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.28 | 0.07 | Thymidine kinase |
| 2qpo.1.D | 15.15 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.28 | 0.07 | Thymidine kinase |
| 2qpo.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.28 | 0.07 | Thymidine kinase |
| 5bul.1.A | 18.18 | monomer | HHblits | X-ray | 1.98Å | 0.28 | 0.07 | flavin-dependent halogenase triple mutant |
| 5bva.1.A | 18.18 | monomer | HHblits | X-ray | 1.87Å | 0.28 | 0.07 | flavin-dependent halogenase |
| 4qi7.1.A | 27.27 | monomer | HHblits | X-ray | 2.90Å | 0.28 | 0.07 | Cellobiose dehydrogenase |
| 4qi7.2.A | 27.27 | monomer | HHblits | X-ray | 2.90Å | 0.28 | 0.07 | Cellobiose dehydrogenase |
| 2dc1.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | L-aspartate dehydrogenase |
| 2dc1.1.B | 25.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | L-aspartate dehydrogenase |
| 1dxl.1.A | 11.76 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1dxl.1.B | 11.76 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1dxl.2.A | 11.76 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1dxl.2.B | 11.76 | homo-dimer | HHblits | X-ray | 3.15Å | 0.26 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 5tue.1.A | 14.71 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | Tetracycline destructase Tet(50) |
| 5tue.2.A | 14.71 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.08 | Tetracycline destructase Tet(50) |
| 5tui.2.A | 14.71 | monomer | HHblits | X-ray | 1.75Å | 0.26 | 0.08 | Tetracycline destructase Tet(50) |
| 3bgd.1.A | 14.71 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.08 | Thiopurine S-methyltransferase |
| 3bgi.1.A | 14.71 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.08 | Thiopurine S-methyltransferase |
| 3bgi.2.A | 14.71 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.08 | Thiopurine S-methyltransferase |
| 2gb4.1.A | 14.71 | monomer | HHblits | X-ray | 1.25Å | 0.26 | 0.08 | Thiopurine S-methyltransferase |
| 1i3k.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.50Å | 0.24 | 0.08 | UDP-GLUCOSE 4-EPIMERASE |
| 1hzj.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.50Å | 0.24 | 0.08 | UDP-GALACTOSE 4-EPIMERASE |
| 1hzj.1.B | 20.00 | homo-dimer | HHblits | X-ray | 1.50Å | 0.24 | 0.08 | UDP-GALACTOSE 4-EPIMERASE |
| 3ea0.1.A | 8.57 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.08 | ATPase, ParA family |
| 3ea0.1.B | 8.57 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.08 | ATPase, ParA family |
| 1xjc.1.A | 14.29 | monomer | HHblits | X-ray | 2.10Å | 0.24 | 0.08 | MobB protein homolog |
| 5vr8.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | UDP-glucose 6-dehydrogenase |
| 3khu.1.A | 18.18 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | UDP-glucose 6-dehydrogenase |
| 2qg4.1.B | 18.18 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.28 | 0.07 | UDP-glucose 6-dehydrogenase |
| 2qg4.1.A | 18.18 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.28 | 0.07 | UDP-glucose 6-dehydrogenase |
| 3ay3.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.07 | NAD-dependent epimerase/dehydratase |
| 2cul.1.A | 24.24 | monomer | HHblits | X-ray | 1.65Å | 0.28 | 0.07 | Glucose-inhibited division protein A-related protein, probable oxidoreductase |
| 1edo.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | BETA-KETO ACYL CARRIER PROTEIN REDUCTASE |
| 5buk.1.A | 21.21 | monomer | HHblits | X-ray | 1.95Å | 0.28 | 0.07 | FADH2-dependent halogenase |
| 3ljp.1.A | 24.24 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.07 | Choline oxidase |
| 3nne.1.A | 24.24 | monomer | HHblits | X-ray | 2.47Å | 0.28 | 0.07 | Choline oxidase |
| 1uf9.1.A | 32.26 | monomer | HHblits | X-ray | 2.80Å | 0.33 | 0.07 | TT1252 protein |
| 1uf9.2.A | 32.26 | monomer | HHblits | X-ray | 2.80Å | 0.33 | 0.07 | TT1252 protein |
| 1uf9.3.A | 32.26 | monomer | HHblits | X-ray | 2.80Å | 0.33 | 0.07 | TT1252 protein |
| 1zoy.1.A | 28.13 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.31 | 0.07 | FAD-binding protein |
| 2h88.3.A | 28.13 | hetero-oligomer | HHblits | X-ray | 1.74Å | 0.31 | 0.07 | SUCCINATE DEHYDROGENASE FLAVOPROTEIN SUBUNIT |
| 4ytp.1.A | 28.13 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.31 | 0.07 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial |
| 5x62.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.31 | 0.07 | Carnosine N-methyltransferase |
| 2qmo.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.47Å | 0.31 | 0.07 | Dethiobiotin synthetase |
| 3qj4.1.A | 20.59 | monomer | HHblits | X-ray | 2.50Å | 0.26 | 0.08 | Renalase |
| 3qj4.2.A | 20.59 | monomer | HHblits | X-ray | 2.50Å | 0.26 | 0.08 | Renalase |
| 5l3q.1.B | 14.71 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.26 | 0.08 | Signal recognition particle receptor subunit alpha |
| 5mbx.1.A | 14.71 | monomer | HHblits | X-ray | 1.40Å | 0.26 | 0.08 | Peroxisomal N(1)-acetyl-spermine/spermidine oxidase |
| 2e1m.1.A | 21.21 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.07 | L-glutamate oxidase |
| 1kdg.1.A | 27.27 | homo-dimer | HHblits | X-ray | 1.50Å | 0.28 | 0.07 | cellobiose dehydrogenase |
| 4bmv.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.08 | SHORT-CHAIN DEHYDROGENASE |
| 4usr.1.A | 14.71 | monomer | HHblits | X-ray | 1.83Å | 0.26 | 0.08 | MONOOXYGENASE |
| 3ics.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.94Å | 0.26 | 0.08 | Coenzyme A-Disulfide Reductase |
| 1zy8.1.A | 11.76 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 1zy8.5.A | 11.76 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 1zy8.5.B | 11.76 | hetero-oligomer | HHblits | X-ray | 2.59Å | 0.26 | 0.08 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 4yrb.1.A | 17.65 | homo-dimer | HHblits | X-ray | 3.25Å | 0.26 | 0.08 | L-threonine 3-dehydrogenase, mitochondrial |
| 3nks.1.A | 14.71 | monomer | HHblits | X-ray | 1.90Å | 0.26 | 0.08 | Protoporphyrinogen oxidase |
| 1gnd.1.A | 14.71 | monomer | HHblits | X-ray | 1.81Å | 0.26 | 0.08 | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR |
| 1d5t.1.A | 14.71 | monomer | HHblits | X-ray | 1.04Å | 0.26 | 0.08 | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR |
| 1lv0.1.A | 14.71 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.08 | RAB GDP disossociation inhibitor alpha |
| 5u4q.1.A | 28.13 | homo-dimer | HHblits | X-ray | 1.50Å | 0.30 | 0.07 | dTDP-glucose 4,6-dehydratase |
| 1xjc.1.A | 21.88 | monomer | HHblits | X-ray | 2.10Å | 0.30 | 0.07 | MobB protein homolog |
| 5twb.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.82Å | 0.30 | 0.07 | Ferredoxin--NADP reductase |
| 5twc.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.31Å | 0.30 | 0.07 | Ferredoxin--NADP reductase |
| 3ake.1.A | 25.00 | monomer | HHblits | X-ray | 1.50Å | 0.30 | 0.07 | Cytidylate kinase |
| 3akc.1.A | 25.00 | monomer | HHblits | X-ray | 1.65Å | 0.30 | 0.07 | Cytidylate kinase |
| 3w8n.1.A | 25.00 | monomer | HHblits | X-ray | 2.20Å | 0.30 | 0.07 | Cytidylate kinase |
| 3w90.1.A | 25.00 | monomer | HHblits | X-ray | 1.65Å | 0.30 | 0.07 | Cytidylate kinase |
| 4hb9.1.A | 18.18 | monomer | HHblits | X-ray | 1.93Å | 0.28 | 0.07 | Similarities with probable monooxygenase |
| 3kjg.2.A | 15.15 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kjg.1.A | 15.15 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kji.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.13Å | 0.28 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kji.2.A | 15.15 | homo-dimer | HHblits | X-ray | 2.13Å | 0.28 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kjh.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kje.1.A | 15.15 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 4c3x.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | 3-KETOSTEROID DEHYDROGENASE |
| 2ofp.1.A | 24.24 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | Ketopantoate reductase |
| 2ofp.2.A | 24.24 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | Ketopantoate reductase |
| 1ks9.1.A | 24.24 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.07 | 2-DEHYDROPANTOATE 2-REDUCTASE |
| 3of5.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.52Å | 0.28 | 0.07 | Dethiobiotin synthetase |
| 3fg2.1.A | 18.18 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.07 | Putative rubredoxin reductase |
| 2eq8.1.A | 18.18 | hetero-oligomer | HHblits | X-ray | 1.94Å | 0.28 | 0.07 | Pyruvate dehydrogenase complex, dihydrolipoamide dehydrogenase E3 component |
| 1bhy.1.A | 21.21 | homo-dimer | HHblits | X-ray | 4.18Å | 0.28 | 0.07 | P64K |
| 1ojt.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.75Å | 0.28 | 0.07 | SURFACE PROTEIN |
| 5evy.1.A | 21.21 | monomer | HHblits | X-ray | 2.47Å | 0.28 | 0.07 | Salicylate hydroxylase |
| 2gpw.1.A | 5.71 | monomer | HHblits | X-ray | 2.20Å | 0.23 | 0.08 | Biotin carboxylase |
| 2gpw.4.A | 5.71 | monomer | HHblits | X-ray | 2.20Å | 0.23 | 0.08 | Biotin carboxylase |
| 4yra.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.08 | L-threonine 3-dehydrogenase, mitochondrial |
| 4yr9.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.08 | L-threonine 3-dehydrogenase, mitochondrial |
| 4yra.1.B | 17.65 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.08 | L-threonine 3-dehydrogenase, mitochondrial |
| 3ic9.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.08 | dihydrolipoamide dehydrogenase |
| 3ic9.1.B | 17.65 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.08 | dihydrolipoamide dehydrogenase |
| 3ic9.2.B | 17.65 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.08 | dihydrolipoamide dehydrogenase |
| 4wop.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.39Å | 0.30 | 0.07 | ATP-dependent dethiobiotin synthetase BioD |
| 3fmi.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.18Å | 0.30 | 0.07 | Dethiobiotin synthetase |
| 3fmi.1.B | 21.88 | homo-dimer | HHblits | X-ray | 2.18Å | 0.30 | 0.07 | Dethiobiotin synthetase |
| 3fmf.2.A | 21.88 | homo-dimer | HHblits | X-ray | 2.05Å | 0.30 | 0.07 | Dethiobiotin synthetase |
| 1nks.1.C | 21.88 | homo-trimer | HHblits | X-ray | 2.57Å | 0.30 | 0.07 | ADENYLATE KINASE |
| 1nks.1.B | 21.88 | homo-trimer | HHblits | X-ray | 2.57Å | 0.30 | 0.07 | ADENYLATE KINASE |
| 1nks.1.A | 21.88 | homo-trimer | HHblits | X-ray | 2.57Å | 0.30 | 0.07 | ADENYLATE KINASE |
| 2foi.1.A | 15.15 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.07 | enoyl-acyl carrier reductase |
| 5j7x.1.A | 21.21 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.07 | Dimethylaniline monooxygenase, putative |
| 5l46.1.A | 21.21 | monomer | HHblits | X-ray | 3.09Å | 0.28 | 0.07 | Dimethylglycine dehydrogenase, mitochondrial |
| 4p9s.1.A | 21.21 | monomer | HHblits | X-ray | 2.32Å | 0.28 | 0.07 | Dimethylglycine dehydrogenase |
| 3iim.1.A | 21.21 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | Coilin-interacting nuclear ATPase protein |
| 1jax.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | conserved hypothetical protein |
| 1jay.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.07 | Coenzyme F420H2:NADP+ Oxidoreductase (FNO) |
| 4tlz.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.41Å | 0.27 | 0.07 | KtzI |
| 1v59.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.07 | Dihydrolipoamide dehydrogenase |
| 1jeh.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1jeh.1.B | 15.15 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 4fk1.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | Putative thioredoxin reductase |
| 4z2t.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.27 | 0.07 | 2-hydroxybiphenyl-3-monooxygenase |
| 3axb.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.92Å | 0.27 | 0.07 | Putative oxidoreductase |
| 3vqr.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.01Å | 0.27 | 0.07 | Putative oxidoreductase |
| 5thk.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.40Å | 0.25 | 0.08 | Putative dehydrogenase |
| 4cnj.1.A | 14.71 | monomer | HHblits | X-ray | 2.70Å | 0.25 | 0.08 | L-AMINO ACID OXIDASE |
| 4cnj.4.A | 14.71 | monomer | HHblits | X-ray | 2.70Å | 0.25 | 0.08 | L-AMINO ACID OXIDASE |
| 1bhy.1.A | 11.76 | homo-dimer | HHblits | X-ray | 4.18Å | 0.25 | 0.08 | P64K |
| 1ojt.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.75Å | 0.25 | 0.08 | SURFACE PROTEIN |
| 5u25.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.08 | Dihydrolipoamide dehydrogenase (E3 component of pyruvate and 2-oxoglutarate dehydrogenase complexes) |
| 3urh.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.08 | Dihydrolipoyl dehydrogenase |
| 3sm8.1.A | 25.00 | monomer | HHblits | X-ray | 1.07Å | 0.30 | 0.07 | FAD-dependent catabolic D-arginine dehydrogenase, DauA |
| 1z83.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.07 | Adenylate kinase 1 |
| 2c95.1.A | 18.75 | monomer | HHblits | X-ray | 1.71Å | 0.30 | 0.07 | ADENYLATE KINASE 1 |
| 3d1c.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.07 | Flavin-containing Putative Monooxygenase |
| 2phh.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1phh.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 1pxb.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.07 | P-HYDROXYBENZOATE HYDROXYLASE |
| 5jzv.1.A | 21.21 | monomer | HHblits | X-ray | 2.07Å | 0.27 | 0.07 | Adenylate kinase isoenzyme 6 |
| 2j41.1.B | 18.18 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | GUANYLATE KINASE |
| 2j41.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | GUANYLATE KINASE |
| 4qrh.1.B | 18.18 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.07 | Guanylate kinase |
| 4qrh.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.07 | Guanylate kinase |
| 1qrr.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.07 | sulfolipid biosynthesis (SQD1) PROTEIN |
| 1b8s.1.A | 18.18 | monomer | HHblits | X-ray | 1.65Å | 0.27 | 0.07 | PROTEIN (CHOLESTEROL OXIDASE) |
| 4xwr.1.A | 18.18 | monomer | HHblits | X-ray | 1.10Å | 0.27 | 0.07 | Cholesterol oxidase |
| 3gyj.1.A | 18.18 | monomer | HHblits | X-ray | 0.92Å | 0.27 | 0.07 | Cholesterol oxidase |
| 3cp8.1.A | 21.21 | homo-dimer | HHblits | X-ray | 3.20Å | 0.27 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA |
| 3cp8.2.A | 21.21 | homo-dimer | HHblits | X-ray | 3.20Å | 0.27 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA |
| 3p1w.1.A | 14.71 | monomer | HHblits | X-ray | 1.85Å | 0.25 | 0.08 | RabGDI protein |
| 5jwa.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.16Å | 0.25 | 0.08 | NADH dehydrogenase, putative |
| 3e1t.1.A | 17.65 | monomer | HHblits | X-ray | 2.05Å | 0.25 | 0.08 | Halogenase |
| 1v0j.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.25Å | 0.25 | 0.08 | UDP-GALACTOPYRANOSE MUTASE |
| 2dki.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.50Å | 0.25 | 0.08 | 3-HYDROXYBENZOATE HYDROXYLASE |
| 2oa1.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.08 | Tryptophan halogenase |
| 3l8k.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.07 | Dihydrolipoyl dehydrogenase |
| 4hut.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.95Å | 0.30 | 0.07 | Cob(I)yrinic acid a,c-diamide adenosyltransferase |
| 4hut.1.B | 21.88 | homo-dimer | HHblits | X-ray | 1.95Å | 0.30 | 0.07 | Cob(I)yrinic acid a,c-diamide adenosyltransferase |
| 3v9p.1.A | 25.81 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.07 | Thymidylate kinase |
| 5tul.1.A | 15.15 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | Tetracycline destructase Tet(55) |
| 1y56.1.A | 21.21 | hetero-oligomer | HHblits | X-ray | 2.86Å | 0.27 | 0.07 | hypothetical protein PH1363 |
| 3gsi.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | N,N-dimethylglycine oxidase |
| 4mif.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | Pyranose 2-oxidase |
| 3k7t.1.A | 24.24 | homo-dimer | HHblits | X-ray | 2.85Å | 0.27 | 0.07 | 6-hydroxy-L-nicotine oxidase |
| 3k7m.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.95Å | 0.27 | 0.07 | 6-hydroxy-L-nicotine oxidase |
| 3hzl.1.A | 21.21 | monomer | HHblits | X-ray | 1.55Å | 0.27 | 0.07 | NikD protein |
| 2q6u.1.A | 21.21 | monomer | HHblits | X-ray | 1.75Å | 0.27 | 0.07 | NikD protein |
| 2olo.1.A | 21.21 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | nikD protein |
| 2oln.1.A | 21.21 | monomer | HHblits | X-ray | 1.15Å | 0.27 | 0.07 | nikD protein |
| 4r01.1.A | 8.82 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.25 | 0.08 | putative NADH-flavin reductase |
| 4r01.1.B | 8.82 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.25 | 0.08 | putative NADH-flavin reductase |
| 5xb5.1.A | 40.00 | homo-dimer | HHblits | X-ray | 2.23Å | 0.34 | 0.07 | Thymidylate kinase |
| 3vr8.1.A | 21.88 | hetero-oligomer | HHblits | X-ray | 2.81Å | 0.29 | 0.07 | Flavoprotein subunit of complex II |
| 4ysx.1.A | 21.88 | hetero-oligomer | HHblits | X-ray | 2.25Å | 0.29 | 0.07 | Succinate dehydrogenase flavoprotein |
| 1y63.1.A | 18.75 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.07 | Lmaj004144AAA protein |
| 2gps.1.A | 2.86 | monomer | HHblits | X-ray | 2.80Å | 0.22 | 0.08 | Biotin carboxylase |
| 4ige.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.07 | Enoyl-acyl carrier reductase |
| 3hyv.1.A | 18.18 | homo-trimer | HHblits | X-ray | 2.30Å | 0.27 | 0.07 | Sulfide-quinone reductase |
| 4quk.1.A | 15.15 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | Dihydroflavonol-4-reductase |
| 1uky.1.A | 15.15 | monomer | HHblits | X-ray | 2.13Å | 0.27 | 0.07 | URIDYLATE KINASE |
| 3iwa.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.07 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 3h8l.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.57Å | 0.27 | 0.07 | NADH oxidase |
| 3o0h.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | Glutathione reductase |
| 3n2i.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.07 | Thymidylate kinase |
| 2wes.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.07 | TRYPTOPHAN 5-HALOGENASE |
| 2wet.2.A | 18.18 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | TRYPTOPHAN 5-HALOGENASE |
| 2wet.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | TRYPTOPHAN 5-HALOGENASE |
| 4g9k.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.08 | Rotenone-insensitive NADH-ubiquinone oxidoreductase |
| 4g6g.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.39Å | 0.25 | 0.08 | Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial |
| 1ebd.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.25 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1ebd.1.B | 14.71 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.25 | 0.08 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 3cph.2.A | 14.71 | monomer | HHblits | X-ray | 2.90Å | 0.25 | 0.08 | Rab GDP-dissociation inhibitor |
| 3cph.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.25 | 0.08 | Rab GDP-dissociation inhibitor |
| 1ukv.1.A | 14.71 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.25 | 0.08 | Secretory pathway GDP dissociation inhibitor |
| 1dah.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.64Å | 0.29 | 0.07 | DETHIOBIOTIN SYNTHETASE |
| 1byi.1.A | 21.88 | homo-dimer | HHblits | X-ray | 0.97Å | 0.29 | 0.07 | DETHIOBIOTIN SYNTHASE |
| 1dts.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.65Å | 0.29 | 0.07 | DETHIOBIOTIN SYNTHETASE |
| 2shk.1.A | 22.58 | homo-dimer | HHblits | X-ray | 2.60Å | 0.32 | 0.07 | SHIKIMATE KINASE |
| 2shk.1.B | 22.58 | homo-dimer | HHblits | X-ray | 2.60Å | 0.32 | 0.07 | SHIKIMATE KINASE |
| 4b63.1.A | 8.57 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.22 | 0.08 | L-ORNITHINE N5 MONOOXYGENASE |
| 4ntc.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | GliT |
| 4x9m.1.A | 21.21 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | L-alpha-glycerophosphate oxidase |
| 5fs8.1.A | 12.12 | monomer | HHblits | X-ray | 1.40Å | 0.27 | 0.07 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 4lii.1.A | 12.12 | monomer | HHblits | X-ray | 1.88Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 5fmh.1.A | 12.12 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5kvi.1.A | 12.12 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 1gv4.1.A | 12.12 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | PROGRAMED CELL DEATH PROTEIN 8 |
| 4bur.1.A | 12.12 | monomer | HHblits | X-ray | 2.88Å | 0.27 | 0.07 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 4bur.3.A | 12.12 | monomer | HHblits | X-ray | 2.88Å | 0.27 | 0.07 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 5kvh.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.27Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 1m6i.1.A | 12.12 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | Programmed cell death protein 8 |
| 4fdc.1.A | 12.12 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd4.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.24Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd4.1.B | 12.12 | homo-dimer | HHblits | X-ray | 2.24Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.1.A | 12.12 | monomer | HHblits | X-ray | 2.95Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.2.A | 12.12 | monomer | HHblits | X-ray | 2.95Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.4.A | 12.12 | monomer | HHblits | X-ray | 2.95Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 5miu.1.A | 12.12 | monomer | HHblits | X-ray | 3.50Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.A | 12.12 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.B | 12.12 | homo-dimer | HHblits | X-ray | 3.10Å | 0.27 | 0.07 | Apoptosis-inducing factor 1, mitochondrial |
| 4mo2.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | UDP-galactopyranose mutase |
| 4ywo.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.62Å | 0.27 | 0.07 | Mercuric reductase |
| 5ks8.1.A | 8.82 | hetero-oligomer | HHblits | X-ray | 3.01Å | 0.24 | 0.08 | Pyruvate carboxylase subunit alpha |
| 3qfa.1.A | 20.59 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.24 | 0.08 | Thioredoxin reductase 1, cytoplasmic |
| 3on3.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.19Å | 0.29 | 0.07 | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit |
| 3on3.1.B | 21.88 | homo-dimer | HHblits | X-ray | 2.19Å | 0.29 | 0.07 | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit |
| 1dnc.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.07 | GLUTATHIONE REDUCTASE |
| 2gh5.1.B | 21.88 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.07 | glutathione reductase, mitochondrial |
| 2aaq.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.07 | glutathione reductase |
| 3djg.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Glutathione reductase |
| 2jbv.1.A | 25.00 | monomer | HHblits | X-ray | 1.86Å | 0.29 | 0.07 | CHOLINE OXIDASE |
| 5kvc.1.A | 32.26 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.31 | 0.07 | Halohydrin dehalogenase |
| 2zxi.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG |
| 2zxh.1.A | 15.15 | homo-dimer | HHblits | X-ray | 3.20Å | 0.26 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG |
| 3nta.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.01Å | 0.26 | 0.07 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 3ntd.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.99Å | 0.26 | 0.07 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 4ocg.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.75Å | 0.26 | 0.07 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 3nt6.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 2o3j.1.A | 15.15 | homo-hexamer | HHblits | X-ray | 1.88Å | 0.26 | 0.07 | UDP-glucose 6-dehydrogenase |
| 3dme.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.07 | conserved exported protein |
| 3urh.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase |
| 3sc6.3.A | 15.15 | monomer | HHblits | X-ray | 2.65Å | 0.26 | 0.07 | dTDP-4-dehydrorhamnose reductase |
| 3sc6.2.A | 15.15 | monomer | HHblits | X-ray | 2.65Å | 0.26 | 0.07 | dTDP-4-dehydrorhamnose reductase |
| 3sc6.1.A | 15.15 | monomer | HHblits | X-ray | 2.65Å | 0.26 | 0.07 | dTDP-4-dehydrorhamnose reductase |
| 3ii4.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.42Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase |
| 4m52.1.B | 15.15 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase |
| 3ic9.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.07 | dihydrolipoamide dehydrogenase |
| 3ic9.1.B | 15.15 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.07 | dihydrolipoamide dehydrogenase |
| 3ic9.2.B | 15.15 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.07 | dihydrolipoamide dehydrogenase |
| 2bra.1.A | 21.21 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS |
| 2bra.2.A | 21.21 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS |
| 4txk.1.A | 21.21 | monomer | HHblits | X-ray | 2.88Å | 0.26 | 0.07 | Protein-methionine sulfoxide oxidase MICAL1 |
| 4txi.1.A | 21.21 | monomer | HHblits | X-ray | 2.31Å | 0.26 | 0.07 | Protein-methionine sulfoxide oxidase MICAL1 |
| 1mo9.1.A | 8.82 | homo-dimer | HHblits | X-ray | 1.65Å | 0.24 | 0.08 | orf3 |
| 1mok.1.A | 8.82 | homo-dimer | HHblits | X-ray | 2.80Å | 0.24 | 0.08 | orf3 |
| 2we7.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.90Å | 0.24 | 0.08 | XANTHINE DEHYDROGENASE |
| 2we7.2.A | 14.71 | homo-dimer | HHblits | X-ray | 2.90Å | 0.24 | 0.08 | XANTHINE DEHYDROGENASE |
| 2y0c.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.75Å | 0.29 | 0.07 | UDP-GLUCOSE DEHYDROGENASE |
| 2y0c.2.A | 18.75 | homo-dimer | HHblits | X-ray | 1.75Å | 0.29 | 0.07 | UDP-GLUCOSE DEHYDROGENASE |
| 3fbs.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.15Å | 0.29 | 0.07 | Oxidoreductase |
| 3grt.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | GLUTATHIONE REDUCTASE |
| 1grt.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | GLUTATHIONE REDUCTASE |
| 6b4o.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.73Å | 0.29 | 0.07 | Glutathione reductase |
| 4nkr.1.A | 30.00 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.2.A | 30.00 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.2.B | 30.00 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.3.A | 30.00 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.3.B | 30.00 | homo-dimer | HHblits | X-ray | 2.41Å | 0.34 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1knx.1.A | 24.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.B | 24.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.C | 24.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.D | 24.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.E | 24.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.F | 24.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 4n9x.1.A | 18.18 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Putative monooxygenase |
| 1ek5.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1ek6.1.B | 24.24 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1ek6.1.A | 24.24 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 3r9u.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.36Å | 0.26 | 0.07 | Thioredoxin reductase |
| 3rnm.1.A | 12.12 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase, mitochondrial |
| 4eqw.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.07 | Coenzyme A disulfide reductase |
| 1pjz.1.A | 15.15 | monomer | HHblits | NMR | NA | 0.26 | 0.07 | Thiopurine S-methyltransferase |
| 4dna.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.07 | Probable glutathione reductase |
| 4dna.1.B | 15.15 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.07 | Probable glutathione reductase |
| 1hyu.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | ALKYL HYDROPEROXIDE REDUCTASE SUBUNIT F |
| 3on5.1.A | 11.76 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.24 | 0.08 | BH1974 protein |
| 3on5.1.B | 11.76 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.24 | 0.08 | BH1974 protein |
| 4m9u.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.60Å | 0.24 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 4ma0.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.98Å | 0.24 | 0.08 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 5kmq.1.A | 8.82 | homo-dimer | HHblits | X-ray | 2.70Å | 0.24 | 0.08 | FAD-dependent pyridine nucleotide-disulfide oxidoreductase |
| 1nmy.1.A | 21.88 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | similar to THYMIDYLATE KINASE (DTMP KINASE) |
| 4ntd.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | Thioredoxin reductase |
| 3pid.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.40Å | 0.29 | 0.07 | UDP-glucose 6-dehydrogenase |
| 3plr.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.07 | UDP-glucose 6-dehydrogenase |
| 3pjg.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.70Å | 0.29 | 0.07 | UDP-glucose 6-dehydrogenase |
| 3phl.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | UDP-glucose 6-dehydrogenase |
| 4qi4.1.A | 28.13 | monomer | HHblits | X-ray | 2.70Å | 0.29 | 0.07 | Cellobiose dehydrogenase |
| 1naa.1.A | 28.13 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Cellobiose dehydrogenase |
| 2e5v.1.A | 19.35 | monomer | HHblits | X-ray | 2.09Å | 0.31 | 0.07 | L-aspartate oxidase |
| 5u9c.1.A | 15.15 | homo-hexamer | HHblits | X-ray | 1.90Å | 0.26 | 0.07 | dTDP-4-dehydrorhamnose Reductase |
| 5u9c.1.E | 15.15 | homo-hexamer | HHblits | X-ray | 1.90Å | 0.26 | 0.07 | dTDP-4-dehydrorhamnose Reductase |
| 4zn0.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | Thioredoxin reductase |
| 4zn0.1.B | 12.12 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | Thioredoxin reductase |
| 1lvl.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.45Å | 0.26 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1ebd.1.A | 18.18 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1ebd.1.B | 18.18 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 3q9t.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.07 | Choline dehydrogenase and related flavoproteins |
| 4d7e.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.08 | L-LYS MONOOXYGENASE |
| 4d7e.1.B | 11.76 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.08 | L-LYS MONOOXYGENASE |
| 4d7e.2.A | 11.76 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.08 | L-LYS MONOOXYGENASE |
| 4d7e.2.B | 11.76 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.08 | L-LYS MONOOXYGENASE |
| 1dnc.1.A | 14.71 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.08 | GLUTATHIONE REDUCTASE |
| 2gh5.1.B | 14.71 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.08 | glutathione reductase, mitochondrial |
| 1grt.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.30Å | 0.24 | 0.08 | GLUTATHIONE REDUCTASE |
| 5b12.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.72Å | 0.28 | 0.07 | Halohydrin epoxidase B |
| 4zd6.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.28 | 0.07 | Halohydrin epoxidase B |
| 4zd6.1.B | 18.75 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.28 | 0.07 | Halohydrin epoxidase B |
| 1i2c.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.60Å | 0.28 | 0.07 | SULFOLIPID BIOSYNTHESIS PROTEIN SQD1 |
| 3guy.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.28 | 0.07 | Short-chain dehydrogenase/reductase SDR |
| 3we0.1.A | 21.88 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.28 | 0.07 | L-amino acid oxidase/monooxygenase |
| 3fim.1.A | 25.00 | monomer | HHblits | X-ray | 2.55Å | 0.28 | 0.07 | Aryl-alcohol oxidase |
| 4eun.1.A | 23.33 | homo-dimer | HHblits | X-ray | 1.60Å | 0.33 | 0.07 | thermoresistant glucokinase |
| 2jas.1.A | 16.13 | homo-dimer | HHblits | X-ray | 2.70Å | 0.31 | 0.07 | DEOXYGUANOSINE KINASE |
| 5tum.1.A | 11.76 | monomer | HHblits | X-ray | 3.30Å | 0.24 | 0.08 | tetracycline destructase Tet(56) |
| 5tum.2.A | 11.76 | monomer | HHblits | X-ray | 3.30Å | 0.24 | 0.08 | tetracycline destructase Tet(56) |
| 4x4j.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.65Å | 0.26 | 0.07 | Putative oxygenase |
| 5b6k.1.A | 18.18 | monomer | HHblits | X-ray | 1.70Å | 0.26 | 0.07 | Uncharacterized protein CgKR1 |
| 1r18.1.A | 12.12 | monomer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | Protein-L-isoaspartate(D-aspartate)-O-methyltransferase |
| 3da1.1.A | 15.15 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.07 | Glycerol-3-phosphate dehydrogenase |
| 5f3r.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.04Å | 0.26 | 0.07 | UDP-galactopyranose mutase |
| 3oz2.1.A | 18.18 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.07 | Digeranylgeranylglycerophospholipid reductase |
| 2rgo.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Alpha-Glycerophosphate Oxidase |
| 2rgh.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.07 | Alpha-Glycerophosphate Oxidase |
| 5lv9.1.A | 15.15 | monomer | HHblits | X-ray | 2.33Å | 0.26 | 0.07 | thermophilic tryptophan halogenase |
| 4ha6.1.A | 21.21 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.07 | Pyridoxine 4-oxidase |
| 3t37.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.19Å | 0.26 | 0.07 | Probable dehydrogenase |
| 5vt3.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.98Å | 0.26 | 0.07 | Thioredoxin reductase |
| 1zmo.1.A | 15.63 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | halohydrin dehalogenase |
| 1yjq.1.A | 25.00 | monomer | HHblits | X-ray | 2.09Å | 0.28 | 0.07 | 2-dehydropantoate 2-reductase |
| 2yqu.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.07 | 2-oxoglutarate dehydrogenase E3 component |
| 2eq7.1.A | 18.75 | hetero-oligomer | HHblits | X-ray | 1.80Å | 0.28 | 0.07 | 2-oxoglutarate dehydrogenase E3 component |
| 5u1o.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.31Å | 0.28 | 0.07 | Glutathione reductase |
| 5uwy.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.72Å | 0.28 | 0.07 | Thioredoxin reductase |
| 3k4c.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 2f6c.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.84Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 3bg7.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.07 | Pyranose oxidase |
| 2igk.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.07 | Pyranose oxidase |
| 1tt0.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.28 | 0.07 | pyranose oxidase |
| 3k4j.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 3fdy.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.55Å | 0.28 | 0.07 | Pyranose oxidase (Pyranose 2-oxidase) |
| 4mog.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 4moh.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 2igo.2.B | 28.13 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.28 | 0.07 | Pyranose oxidase |
| 3lsm.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 3pl8.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.35Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 2f5v.1.A | 28.13 | homo-tetramer | HHblits | X-ray | 1.41Å | 0.28 | 0.07 | Pyranose 2-oxidase |
| 5tuk.1.A | 11.76 | monomer | HHblits | X-ray | 1.85Å | 0.23 | 0.08 | Tetracycline destructase Tet(51) |
| 5tuk.2.A | 11.76 | monomer | HHblits | X-ray | 1.85Å | 0.23 | 0.08 | Tetracycline destructase Tet(51) |
| 5tuk.3.A | 11.76 | monomer | HHblits | X-ray | 1.85Å | 0.23 | 0.08 | Tetracycline destructase Tet(51) |
| 5tuk.4.A | 11.76 | monomer | HHblits | X-ray | 1.85Å | 0.23 | 0.08 | Tetracycline destructase Tet(51) |
| 5tiv.1.A | 15.15 | monomer | HHblits | X-ray | 1.43Å | 0.26 | 0.07 | Sulfotransferase |
| 5tv2.1.A | 9.09 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.07 | Elongation factor G |
| 4pvc.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | NADPH-dependent methylglyoxal reductase GRE2 |
| 4pvd.1.A | 15.15 | monomer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | NADPH-dependent methylglyoxal reductase GRE2 |
| 4pvd.3.A | 15.15 | monomer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | NADPH-dependent methylglyoxal reductase GRE2 |
| 4eip.1.A | 21.21 | monomer | HHblits | X-ray | 2.33Å | 0.26 | 0.07 | Putative FAD-monooxygenase |
| 4eip.2.A | 21.21 | monomer | HHblits | X-ray | 2.33Å | 0.26 | 0.07 | Putative FAD-monooxygenase |
| 4eiq.1.A | 21.21 | monomer | HHblits | X-ray | 2.76Å | 0.26 | 0.07 | Putative FAD-monooxygenase |
| 4eiq.2.A | 21.21 | monomer | HHblits | X-ray | 2.76Å | 0.26 | 0.07 | Putative FAD-monooxygenase |
| 4at0.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.60Å | 0.26 | 0.07 | 3-KETOSTEROID-DELTA4-5ALPHA-DEHYDROGENASE |
| 3c4n.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Uncharacterized protein DR_0571 |
| 5ttk.1.A | 18.18 | monomer | HHblits | X-ray | 2.51Å | 0.26 | 0.07 | Amine oxidase |
| 5ttj.2.A | 18.18 | monomer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | Amine oxidase |
| 4h7u.1.A | 18.18 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.07 | Pyranose dehydrogenase |
| 3etn.1.A | 12.12 | homo-tetramer | HHblits | X-ray | 1.70Å | 0.26 | 0.07 | putative phosphosugar isomerase involved in capsule formation |
| 2q7v.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.07 | Thioredoxin reductase |
| 2q7v.1.B | 12.12 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.07 | Thioredoxin reductase |
| 1mrn.1.A | 22.58 | homo-dimer | HHblits | X-ray | 2.45Å | 0.31 | 0.07 | Thymidylate Kinase |
| 1w2h.1.A | 22.58 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.07 | THYMIDYLATE KINASE TMK |
| 1rkb.1.A | 21.88 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | Protein AD-004 |
| 6aon.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.72Å | 0.28 | 0.07 | Dihydrolipoyl dehydrogenase |
| 2hqm.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.40Å | 0.28 | 0.07 | Glutathione reductase |
| 1xdi.1.A | 21.88 | homo-tetramer | HHblits | X-ray | 2.81Å | 0.28 | 0.07 | Rv3303c-lpdA |
| 5u25.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.07 | Dihydrolipoamide dehydrogenase (E3 component of pyruvate and 2-oxoglutarate dehydrogenase complexes) |
| 2bc0.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.07 | NADH Oxidase |
| 5kow.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.07 | Pentachlorophenol 4-monooxygenase |
| 3pvc.1.A | 18.18 | monomer | HHblits | X-ray | 2.31Å | 0.25 | 0.07 | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC |
| 4xgk.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.07 | UDP-galactopyranose mutase |
| 5br7.1.A | 21.21 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.07 | UDP-galactopyranose mutase |
| 1i8t.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.07 | UDP-GALACTOPYRANOSE MUTASE |
| 1i8t.1.B | 21.21 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.07 | UDP-GALACTOPYRANOSE MUTASE |
| 3if9.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.07 | Glycine oxidase |
| 1ryi.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.25 | 0.07 | GLYCINE OXIDASE |
| 1ng3.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.25 | 0.07 | Glycine oxidase |
| 5kwe.1.A | 33.33 | homo-tetramer | HHblits | X-ray | 1.68Å | 0.33 | 0.07 | Halohydrin dehalogenase |
| 1nks.1.C | 30.00 | homo-trimer | HHblits | X-ray | 2.57Å | 0.33 | 0.07 | ADENYLATE KINASE |
| 1nks.1.B | 30.00 | homo-trimer | HHblits | X-ray | 2.57Å | 0.33 | 0.07 | ADENYLATE KINASE |
| 1nks.1.A | 30.00 | homo-trimer | HHblits | X-ray | 2.57Å | 0.33 | 0.07 | ADENYLATE KINASE |
| 3woh.1.A | 25.81 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.07 | SiaM |
| 4ivn.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.07 | Transcriptional regulator |
| 3ka7.1.A | 21.88 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.07 | Oxidoreductase |
| 1chu.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.07 | PROTEIN (L-ASPARTATE OXIDASE) |
| 1knp.1.A | 25.00 | monomer | HHblits | X-ray | 2.60Å | 0.28 | 0.07 | L-aspartate oxidase |
| 1tev.1.A | 12.50 | monomer | HHblits | X-ray | 2.10Å | 0.28 | 0.07 | UMP-CMP kinase |
| 4mua.1.A | 18.18 | monomer | HHblits | X-ray | 1.95Å | 0.25 | 0.07 | Sulfotransferase |
| 3ab1.1.A | 9.09 | homo-dimer | HHblits | X-ray | 2.39Å | 0.25 | 0.07 | Ferredoxin--NADP reductase |
| 3ab1.1.B | 9.09 | homo-dimer | HHblits | X-ray | 2.39Å | 0.25 | 0.07 | Ferredoxin--NADP reductase |
| 3be4.1.A | 19.35 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.07 | Adenylate kinase |
| 3oc4.1.A | 16.13 | homo-dimer | HHblits | X-ray | 2.60Å | 0.30 | 0.07 | Oxidoreductase, pyridine nucleotide-disulfide family |
| 3oc4.1.B | 16.13 | homo-dimer | HHblits | X-ray | 2.60Å | 0.30 | 0.07 | Oxidoreductase, pyridine nucleotide-disulfide family |
| 2ar7.1.A | 19.35 | monomer | HHblits | X-ray | 2.15Å | 0.30 | 0.07 | Adenylate kinase 4 |
| 2ar7.2.A | 19.35 | monomer | HHblits | X-ray | 2.15Å | 0.30 | 0.07 | Adenylate kinase 4 |
| 2bbw.1.A | 19.35 | monomer | HHblits | X-ray | 2.05Å | 0.30 | 0.07 | adenylate kinase 4, AK4 |
| 2bbw.2.A | 19.35 | monomer | HHblits | X-ray | 2.05Å | 0.30 | 0.07 | adenylate kinase 4, AK4 |
| 4j56.1.A | 29.03 | hetero-oligomer | HHblits | X-ray | 2.37Å | 0.30 | 0.07 | Thioredoxin reductase 2 |
| 4j57.1.A | 29.03 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.30 | 0.07 | Thioredoxin reductase 2 |
| 4b1b.1.A | 29.03 | homo-dimer | HHblits | X-ray | 2.90Å | 0.30 | 0.07 | THIOREDOXIN REDUCTASE |
| 3hjn.1.A | 33.33 | homo-dimer | HHblits | X-ray | 2.10Å | 0.33 | 0.07 | Thymidylate kinase |
| 3hjn.1.B | 33.33 | homo-dimer | HHblits | X-ray | 2.10Å | 0.33 | 0.07 | Thymidylate kinase |
| 4la1.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.07 | Thioredoxin glutathione reductase |
| 3uwk.1.A | 18.75 | monomer | HHblits | X-ray | 1.91Å | 0.28 | 0.07 | Thymidylate kinase |
| 1cc2.1.A | 18.75 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.07 | PROTEIN (CHOLESTEROL OXIDASE) |
| 5v36.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.88Å | 0.27 | 0.07 | Glutathione reductase |
| 5vqc.1.A | 25.00 | monomer | HHblits | X-ray | 3.32Å | 0.27 | 0.07 | Rifampin monooxygenase |
| 2qae.1.A | 9.09 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.07 | Dihydrolipoyl dehydrogenase |
| 5er0.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.41Å | 0.25 | 0.07 | NADH oxidase |
| 5voh.1.A | 12.12 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.25 | 0.07 | NADH oxidase |
| 3a4l.1.A | 22.58 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.07 | L-seryl-tRNA(Sec) kinase |
| 3a4l.1.B | 22.58 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.07 | L-seryl-tRNA(Sec) kinase |
| 3am1.1.C | 22.58 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.07 | L-seryl-tRNA(Sec) kinase |
| 2cdn.1.A | 22.58 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.07 | ADENYLATE KINASE |
| 1cp2.1.A | 30.00 | homo-dimer | HHblits | X-ray | 1.93Å | 0.32 | 0.07 | NITROGENASE IRON PROTEIN |
| 1cp2.1.B | 30.00 | homo-dimer | HHblits | X-ray | 1.93Å | 0.32 | 0.07 | NITROGENASE IRON PROTEIN |
| 4nte.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | DepH |
| 4nte.1.B | 21.88 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | DepH |
| 4jna.1.A | 21.88 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | DepH |
| 4jn9.1.A | 21.88 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | DepH |
| 4jn9.1.B | 21.88 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | DepH |
| 4b4o.1.A | 18.18 | monomer | HHblits | X-ray | 2.70Å | 0.25 | 0.07 | EPIMERASE FAMILY PROTEIN SDR39U1 |
| 3g17.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.07 | Similar to 2-dehydropantoate 2-reductase |
| 3g17.1.B | 12.12 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.07 | Similar to 2-dehydropantoate 2-reductase |
| 4s3m.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.60Å | 0.25 | 0.07 | 2-dehydropantoate 2-reductase |
| 4s3m.1.B | 12.12 | homo-dimer | HHblits | X-ray | 2.60Å | 0.25 | 0.07 | 2-dehydropantoate 2-reductase |
| 3ean.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.75Å | 0.25 | 0.07 | Thioredoxin reductase 1 |
| 1h6v.1.A | 21.21 | homo-dimer | HHblits | X-ray | 3.00Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 2zzc.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.60Å | 0.25 | 0.07 | Thioredoxin reductase 1, cytoplasmic |
| 2zzb.1.B | 21.21 | homo-dimer | HHblits | X-ray | 3.20Å | 0.25 | 0.07 | Thioredoxin reductase 1, cytoplasmic |
| 2cfy.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.70Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE 1 |
| 2j3n.1.A | 21.21 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE 1 |
| 4rpg.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.07 | UDP-galactopyranose mutase |
| 4rpl.2.B | 18.18 | homo-dimer | HHblits | X-ray | 2.25Å | 0.25 | 0.07 | UDP-galactopyranose mutase |
| 4jky.1.A | 22.58 | monomer | HHblits | X-ray | 2.37Å | 0.30 | 0.07 | Adenylate kinase |
| 4jky.2.A | 22.58 | monomer | HHblits | X-ray | 2.37Å | 0.30 | 0.07 | Adenylate kinase |
| 4jl5.1.A | 22.58 | monomer | HHblits | X-ray | 1.24Å | 0.30 | 0.07 | Adenylate kinase |
| 4jl5.2.A | 22.58 | monomer | HHblits | X-ray | 1.24Å | 0.30 | 0.07 | Adenylate kinase |
| 4gmd.1.B | 30.00 | homo-dimer | HHblits | X-ray | 1.98Å | 0.32 | 0.07 | Thymidylate kinase |
| 4gmd.1.A | 30.00 | homo-dimer | HHblits | X-ray | 1.98Å | 0.32 | 0.07 | Thymidylate kinase |
| 4edh.1.A | 30.00 | homo-dimer | HHblits | X-ray | 1.32Å | 0.32 | 0.07 | Thymidylate kinase |
| 4e5u.1.A | 30.00 | homo-dimer | HHblits | X-ray | 1.81Å | 0.32 | 0.07 | Thymidylate kinase |
| 4e5u.1.B | 30.00 | homo-dimer | HHblits | X-ray | 1.81Å | 0.32 | 0.07 | Thymidylate kinase |
| 1p9n.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1p9n.1.B | 18.75 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1np6.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1np6.1.B | 18.75 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 5eer.1.A | 9.38 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.07 | 4-hydroxy-tetrahydrodipicolinate reductase |
| 5ees.1.A | 9.38 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.27 | 0.07 | 4-hydroxy-tetrahydrodipicolinate reductase |
| 3ces.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.41Å | 0.25 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA |
| 3cp2.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.90Å | 0.25 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA |
| 4k2x.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.55Å | 0.25 | 0.07 | Polyketide oxygenase/hydroxylase |
| 1yj8.1.A | 9.09 | homo-dimer | HHblits | X-ray | 2.85Å | 0.25 | 0.07 | glycerol-3-phosphate dehydrogenase |
| 3ept.1.A | 18.18 | monomer | HHblits | X-ray | 2.97Å | 0.25 | 0.07 | RebC |
| 2r0c.1.A | 18.18 | monomer | HHblits | X-ray | 1.80Å | 0.25 | 0.07 | RebC |
| 3c4a.1.A | 12.12 | monomer | HHblits | X-ray | 2.30Å | 0.25 | 0.07 | Probable tryptophan hydroxylase vioD |
| 2a87.1.A | 15.15 | homo-dimer | HHblits | X-ray | 3.00Å | 0.25 | 0.07 | Thioredoxin reductase |
| 2a87.1.B | 15.15 | homo-dimer | HHblits | X-ray | 3.00Å | 0.25 | 0.07 | Thioredoxin reductase |
| 1tde.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 3h0k.1.A | 34.48 | homo-dimer | HHblits | X-ray | 3.25Å | 0.35 | 0.07 | UPF0200 protein SSO1041 |
| 3h0k.1.B | 34.48 | homo-dimer | HHblits | X-ray | 3.25Å | 0.35 | 0.07 | UPF0200 protein SSO1041 |
| 4pzl.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Adenylate kinase |
| 4pzl.1.B | 12.90 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Adenylate kinase |
| 4x8o.1.A | 19.35 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.07 | Adenylate kinase |
| 4x8m.1.A | 19.35 | monomer | HHblits | X-ray | 2.60Å | 0.29 | 0.07 | Adenylate kinase |
| 3hpr.1.A | 19.35 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | Adenylate kinase |
| 4np6.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | Adenylate kinase |
| 4np6.1.B | 19.35 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | Adenylate kinase |
| 4x8h.1.A | 19.35 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Adenylate kinase |
| 4x8l.1.A | 19.35 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.07 | Adenylate kinase |
| 4x8l.2.A | 19.35 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.07 | Adenylate kinase |
| 3mrs.1.A | 36.67 | monomer | HHblits | X-ray | 2.40Å | 0.32 | 0.07 | Shikimate kinase |
| 3nrn.1.A | 15.63 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.07 | uncharacterized protein PF1083 |
| 3dgh.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.27 | 0.07 | Thioredoxin reductase 1, mitochondrial |
| 3k5i.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | Phosphoribosyl-aminoimidazole carboxylase |
| 3k5i.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.07 | Phosphoribosyl-aminoimidazole carboxylase |
| 3k5h.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.07 | Phosphoribosyl-aminoimidazole carboxylase |
| 3k5h.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.07 | Phosphoribosyl-aminoimidazole carboxylase |
| 4jdr.1.A | 18.75 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.07 | Dihydrolipoyl dehydrogenase |
| 4ma5.1.A | 15.15 | homo-dimer | HHblits | X-ray | 1.81Å | 0.24 | 0.07 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 5jqw.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.06Å | 0.24 | 0.07 | N5-carboxyaminoimidazole ribonucleotide synthase |
| 5uth.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.95Å | 0.24 | 0.07 | Thioredoxin reductase |
| 5u63.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.99Å | 0.24 | 0.07 | Thioredoxin reductase |
| 1np6.1.A | 22.58 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1np6.1.B | 22.58 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 1dli.1.A | 29.03 | homo-dimer | HHblits | X-ray | 2.31Å | 0.29 | 0.07 | UDP-GLUCOSE DEHYDROGENASE |
| 1dlj.1.A | 29.03 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | UDP-GLUCOSE DEHYDROGENASE |
| 4w5j.3.A | 19.35 | monomer | HHblits | X-ray | 1.65Å | 0.29 | 0.07 | Adenylate kinase |
| 4ntz.1.A | 19.35 | monomer | HHblits | X-ray | 1.69Å | 0.29 | 0.07 | Adenylate kinase |
| 4nu0.1.A | 19.35 | monomer | HHblits | X-ray | 1.49Å | 0.29 | 0.07 | Adenylate kinase |
| 4nu0.2.A | 19.35 | monomer | HHblits | X-ray | 1.49Å | 0.29 | 0.07 | Adenylate kinase |
| 4w5h.1.A | 19.35 | monomer | HHblits | X-ray | 1.96Å | 0.29 | 0.07 | Adenylate kinase |
| 4w5j.1.A | 19.35 | monomer | HHblits | X-ray | 1.65Å | 0.29 | 0.07 | Adenylate kinase |
| 4a7p.1.A | 15.63 | homo-dimer | HHblits | X-ray | 3.40Å | 0.27 | 0.07 | UDP-GLUCOSE DEHYDROGENASE |
| 2npo.1.A | 15.63 | homo-trimer | HHblits | X-ray | 2.20Å | 0.27 | 0.07 | Acetyltransferase |
| 2v6o.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.07 | THIOREDOXIN GLUTATHIONE REDUCTASE |
| 2x8g.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | THIOREDOXIN GLUTATHIONE REDUCTASE |
| 2x99.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.07 | THIOREDOXIN GLUTATHIONE REDUCTASE |
| 3h4k.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.55Å | 0.27 | 0.07 | Thioredoxin glutathione reductase |
| 4mih.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | Pyranose 2-oxidase |
| 2vvl.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.45Å | 0.24 | 0.07 | MONOAMINE OXIDASE N |
| 3zdn.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.55Å | 0.24 | 0.07 | MONOAMINE OXIDASE N |
| 2vvm.1.A | 12.12 | homo-dimer | HHblits | X-ray | 1.85Å | 0.24 | 0.07 | MONOAMINE OXIDASE N |
| 2vvl.4.B | 12.12 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.24 | 0.07 | MONOAMINE OXIDASE N |
| 1via.1.A | 37.93 | homo-dimer | HHblits | X-ray | 1.57Å | 0.34 | 0.07 | shikimate kinase |
| 3l7n.1.A | 12.50 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.07 | Putative uncharacterized protein |
| 3gaz.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.96Å | 0.26 | 0.07 | Alcohol dehydrogenase superfamily protein |
| 4d7e.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | L-LYS MONOOXYGENASE |
| 4d7e.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | L-LYS MONOOXYGENASE |
| 4d7e.2.A | 15.63 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | L-LYS MONOOXYGENASE |
| 4d7e.2.B | 15.63 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | L-LYS MONOOXYGENASE |
| 4o5q.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | Alkyl hydroperoxide reductase subunit F |
| 4o5u.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.65Å | 0.26 | 0.07 | Alkyl hydroperoxide reductase subunit F |
| 4ykg.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Alkyl hydroperoxide reductase subunit F |
| 4ykf.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Alkyl hydroperoxide reductase subunit F |
| 4xvg.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | Alkyl hydroperoxide reductase subunit F |
| 3dgz.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.25Å | 0.24 | 0.07 | Thioredoxin reductase 2 |
| 1zkq.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.60Å | 0.24 | 0.07 | Thioredoxin reductase 2, mitochondrial |
| 4la1.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.35Å | 0.24 | 0.07 | Thioredoxin glutathione reductase |
| 4k5r.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.07 | Oxygenase |
| 1i24.1.A | 25.81 | homo-dimer | HHblits | X-ray | 1.20Å | 0.29 | 0.07 | SULFOLIPID BIOSYNTHESIS PROTEIN SQD1 |
| 1ka9.1.A | 16.13 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.29 | 0.07 | imidazole glycerol phosphate synthase |
| 1xhc.1.A | 29.03 | monomer | HHblits | X-ray | 2.35Å | 0.29 | 0.07 | NADH oxidase /nitrite reductase |
| 3qfa.1.A | 19.35 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.29 | 0.07 | Thioredoxin reductase 1, cytoplasmic |
| 2j3n.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.80Å | 0.29 | 0.07 | THIOREDOXIN REDUCTASE 1 |
| 2cfy.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.70Å | 0.29 | 0.07 | THIOREDOXIN REDUCTASE 1 |
| 2zzc.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.07 | Thioredoxin reductase 1, cytoplasmic |
| 2zzb.1.B | 19.35 | homo-dimer | HHblits | X-ray | 3.20Å | 0.29 | 0.07 | Thioredoxin reductase 1, cytoplasmic |
| 3ps9.1.A | 15.63 | monomer | HHblits | X-ray | 2.54Å | 0.26 | 0.07 | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC |
| 3awi.1.A | 15.63 | monomer | HHblits | X-ray | 3.00Å | 0.26 | 0.07 | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC |
| 3awi.4.A | 15.63 | monomer | HHblits | X-ray | 3.00Å | 0.26 | 0.07 | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC |
| 3awi.6.A | 15.63 | monomer | HHblits | X-ray | 3.00Å | 0.26 | 0.07 | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC |
| 2a8x.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase |
| 1e1m.1.A | 18.75 | monomer | HHblits | X-ray | 1.85Å | 0.26 | 0.07 | ADRENODOXIN REDUCTASE |
| 1e1l.1.A | 18.75 | monomer | HHblits | X-ray | 2.30Å | 0.26 | 0.07 | ADRENODOXIN REDUCTASE |
| 1e6e.2.A | 18.75 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.26 | 0.07 | NADPH\:ADRENODOXIN OXIDOREDUCTASE |
| 1e6e.1.A | 18.75 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.26 | 0.07 | NADPH\:ADRENODOXIN OXIDOREDUCTASE |
| 2v6o.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.07 | THIOREDOXIN GLUTATHIONE REDUCTASE |
| 3dh9.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.25Å | 0.24 | 0.07 | Thioredoxin reductase 1 |
| 2nvk.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.07 | Thioredoxin Reductase |
| 3fmw.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.89Å | 0.24 | 0.07 | Oxygenase |
| 4zo4.1.A | 12.90 | homo-tetramer | HHblits | X-ray | 2.57Å | 0.29 | 0.07 | Dephospho-CoA kinase |
| 4zo4.1.B | 12.90 | homo-tetramer | HHblits | X-ray | 2.57Å | 0.29 | 0.07 | Dephospho-CoA kinase |
| 5h80.1.A | 2.94 | homo-dimer | HHblits | X-ray | 1.70Å | 0.21 | 0.08 | Carboxylase |
| 1xdb.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 4wza.1.E | 26.67 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 4wza.1.F | 26.67 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 1de0.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1xcp.1.A | 26.67 | homo-dimer | HHblits | X-ray | 3.20Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 1xcp.2.A | 26.67 | homo-dimer | HHblits | X-ray | 3.20Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 1m34.1.E | 26.67 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | Nitrogenase Iron Protein 1 |
| 1m1y.1.E | 26.67 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.31 | 0.07 | nitrogenase IRON protein 1 |
| 1m1y.1.F | 26.67 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.31 | 0.07 | nitrogenase IRON protein 1 |
| 2afi.1.E | 26.67 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 2afh.1.E | 26.67 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 2afh.1.F | 26.67 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 1g5p.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1g5p.1.B | 26.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1g1m.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.25Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1g1m.1.B | 26.67 | homo-dimer | HHblits | X-ray | 2.25Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1fp6.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.15Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1nip.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1nip.1.B | 26.67 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN |
| 1xd9.1.A | 26.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.07 | Nitrogenase iron protein 1 |
| 2c8v.1.A | 26.67 | monomer | HHblits | X-ray | 2.50Å | 0.31 | 0.07 | NITROGENASE IRON PROTEIN 1 |
| 2bc1.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.07 | NADH Oxidase |
| 5tr3.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase |
| 5ez7.1.A | 15.15 | monomer | HHblits | X-ray | 2.40Å | 0.24 | 0.07 | flavoenzyme PA4991 |
| 5o5o.1.A | 30.00 | homo-tetramer | HHblits | X-ray | 3.40Å | 0.31 | 0.07 | RNase adapter protein RapZ |
| 5o5o.1.C | 30.00 | homo-tetramer | HHblits | X-ray | 3.40Å | 0.31 | 0.07 | RNase adapter protein RapZ |
| 2pt5.2.A | 20.00 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | Shikimate kinase |
| 2pt5.1.A | 20.00 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | Shikimate kinase |
| 2pt5.3.A | 20.00 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | Shikimate kinase |
| 2pt5.4.A | 20.00 | monomer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | Shikimate kinase |
| 1lpf.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 5u8u.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.35Å | 0.26 | 0.07 | Dihydrolipoyl dehydrogenase |
| 1onf.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | Glutathione reductase |
| 2q0k.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.07 | Thioredoxin reductase |
| 2q0k.1.B | 15.63 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.07 | Thioredoxin reductase |
| 3ish.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.43Å | 0.26 | 0.07 | Thioredoxin reductase |
| 5vdn.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.55Å | 0.26 | 0.07 | Glutathione oxidoreductase |
| 1ges.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.74Å | 0.26 | 0.07 | GLUTATHIONE REDUCTASE |
| 4eqx.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.70Å | 0.26 | 0.07 | Coenzyme A disulfide reductase |
| 4eqr.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.07 | Coenzyme A disulfide reductase |
| 4emw.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.39Å | 0.26 | 0.07 | Coenzyme A disulfide reductase |
| 4em3.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.98Å | 0.26 | 0.07 | Coenzyme A disulfide reductase |
| 1get.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | GLUTATHIONE REDUCTASE |
| 1yqz.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.54Å | 0.26 | 0.07 | coenzyme A disulfide reductase |
| 4eqs.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.50Å | 0.26 | 0.07 | Coenzyme A disulfide reductase |
| 4ysh.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.23 | 0.07 | Glycine oxidase |
| 4ysh.1.B | 15.15 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.23 | 0.07 | Glycine oxidase |
| 5usx.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | Thioredoxin reductase |
| 5usx.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | Thioredoxin reductase |
| 5utx.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.46Å | 0.26 | 0.07 | Thioredoxin reductase |
| 5utx.1.B | 15.63 | homo-dimer | HHblits | X-ray | 2.46Å | 0.26 | 0.07 | Thioredoxin reductase |
| 3ld9.1.A | 39.29 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.36 | 0.06 | Thymidylate kinase |
| 3dgz.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.25Å | 0.28 | 0.07 | Thioredoxin reductase 2 |
| 1zkq.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.07 | Thioredoxin reductase 2, mitochondrial |
| 1ulz.1.A | 9.09 | homo-dimer | HHblits | X-ray | 2.20Å | 0.23 | 0.07 | pyruvate carboxylase n-terminal domain |
| 2p5u.1.B | 21.88 | homo-dimer | HHblits | X-ray | 2.37Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 2p5u.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.37Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 2p5y.1.A | 21.88 | monomer | HHblits | X-ray | 1.92Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 2p5u.2.B | 21.88 | homo-dimer | HHblits | X-ray | 2.37Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 2h92.1.A | 12.50 | monomer | HHblits | X-ray | 2.30Å | 0.25 | 0.07 | Cytidylate kinase |
| 5o5q.1.A | 30.00 | homo-tetramer | HHblits | X-ray | 3.25Å | 0.30 | 0.07 | RNase adapter protein RapZ |
| 5o5q.1.B | 30.00 | homo-tetramer | HHblits | X-ray | 3.25Å | 0.30 | 0.07 | RNase adapter protein RapZ |
| 5o5q.1.C | 30.00 | homo-tetramer | HHblits | X-ray | 3.25Å | 0.30 | 0.07 | RNase adapter protein RapZ |
| 5o5q.1.D | 30.00 | homo-tetramer | HHblits | X-ray | 3.25Å | 0.30 | 0.07 | RNase adapter protein RapZ |
| 4jlo.1.A | 23.33 | monomer | HHblits | X-ray | 1.73Å | 0.30 | 0.07 | Adenylate kinase |
| 4jlo.2.A | 23.33 | monomer | HHblits | X-ray | 1.73Å | 0.30 | 0.07 | Adenylate kinase |
| 4ike.1.A | 23.33 | monomer | HHblits | X-ray | 1.48Å | 0.30 | 0.07 | Adenylate kinase |
| 2rh5.2.A | 23.33 | monomer | HHblits | X-ray | 2.48Å | 0.30 | 0.07 | Adenylate kinase |
| 2rh5.3.A | 23.33 | monomer | HHblits | X-ray | 2.48Å | 0.30 | 0.07 | Adenylate kinase |
| 2rgx.1.A | 23.33 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.07 | Adenylate kinase |
| 2rh5.1.A | 23.33 | monomer | HHblits | X-ray | 2.48Å | 0.30 | 0.07 | Adenylate kinase |
| 4ike.2.A | 23.33 | monomer | HHblits | X-ray | 1.48Å | 0.30 | 0.07 | Adenylate kinase |
| 3sr0.2.A | 23.33 | monomer | HHblits | X-ray | 1.56Å | 0.30 | 0.07 | Adenylate kinase |
| 4cf7.1.A | 23.33 | monomer | HHblits | X-ray | 1.59Å | 0.30 | 0.07 | ADENYLATE KINASE |
| 4cf7.2.A | 23.33 | monomer | HHblits | X-ray | 1.59Å | 0.30 | 0.07 | ADENYLATE KINASE |
| 3tlx.1.A | 26.67 | monomer | HHblits | X-ray | 2.75Å | 0.30 | 0.07 | Adenylate kinase 2 |
| 3tlx.3.A | 26.67 | monomer | HHblits | X-ray | 2.75Å | 0.30 | 0.07 | Adenylate kinase 2 |
| 3tlx.4.A | 26.67 | monomer | HHblits | X-ray | 2.75Å | 0.30 | 0.07 | Adenylate kinase 2 |
| 4e4y.1.A | 19.35 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.07 | Short chain dehydrogenase family protein |
| 1gy8.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.23 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 3g05.1.A | 12.50 | homo-dimer | HHblits | X-ray | 3.49Å | 0.25 | 0.07 | tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG |
| 2p4h.1.A | 18.75 | monomer | HHblits | X-ray | 1.40Å | 0.25 | 0.07 | Vestitone reductase |
| 1djn.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.07 | TRIMETHYLAMINE DEHYDROGENASE |
| 1djq.1.A | 21.88 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.07 | TRIMETHYLAMINE DEHYDROGENASE |
| 2fja.1.A | 18.75 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.25 | 0.07 | adenylylsulfate reductase, subunit A |
| 1kag.1.A | 24.14 | monomer | HHblits | X-ray | 2.05Å | 0.33 | 0.07 | Shikimate kinase I |
| 1kag.2.A | 24.14 | monomer | HHblits | X-ray | 2.05Å | 0.33 | 0.07 | Shikimate kinase I |
| 5cb6.3.B | 34.48 | homo-dimer | HHblits | X-ray | 2.79Å | 0.33 | 0.07 | Probable adenylyl-sulfate kinase |
| 5cb6.2.B | 34.48 | homo-dimer | HHblits | X-ray | 2.79Å | 0.33 | 0.07 | Probable adenylyl-sulfate kinase |
| 1p4s.1.A | 23.33 | monomer | HHblits | NMR | NA | 0.30 | 0.07 | Adenylate kinase |
| 5eje.1.A | 20.00 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.07 | Adenylate kinase |
| 5m5j.1.A | 9.38 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.07 | Thioredoxin reductase |
| 5m5j.1.B | 9.38 | homo-dimer | HHblits | X-ray | 2.65Å | 0.25 | 0.07 | Thioredoxin reductase |
| 1bxk.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.07 | PROTEIN (DTDP-GLUCOSE 4,6-DEHYDRATASE) |
| 1nhr.1.A | 12.50 | monomer | HHblits | X-ray | 2.10Å | 0.25 | 0.07 | NADH PEROXIDASE |
| 1nhs.1.A | 12.50 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.07 | NADH PEROXIDASE |
| 3c7d.1.A | 12.50 | monomer | HHblits | X-ray | 2.50Å | 0.25 | 0.07 | Octopine dehydrogenase |
| 3c7a.1.A | 12.50 | monomer | HHblits | X-ray | 2.10Å | 0.25 | 0.07 | Octopine dehydrogenase |
| 3iqd.1.A | 12.50 | monomer | HHblits | X-ray | 2.80Å | 0.25 | 0.07 | Octopine dehydrogenase |
| 2osb.1.A | 20.00 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.07 | Adenylate kinase |
| 2osb.2.A | 20.00 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.07 | Adenylate kinase |
| 3hr7.1.A | 37.93 | homo-dimer | HHblits | X-ray | 1.80Å | 0.33 | 0.07 | Shikimate kinase |
| 3hr7.1.B | 37.93 | homo-dimer | HHblits | X-ray | 1.80Å | 0.33 | 0.07 | Shikimate kinase |
| 1zui.1.A | 37.93 | monomer | HHblits | X-ray | 2.30Å | 0.33 | 0.07 | Shikimate kinase |
| 1zuh.1.A | 37.93 | monomer | HHblits | X-ray | 1.80Å | 0.33 | 0.07 | Shikimate kinase |
| 3muf.1.A | 37.93 | monomer | HHblits | X-ray | 2.30Å | 0.33 | 0.07 | Shikimate kinase |
| 3dh9.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.07 | Thioredoxin reductase 1 |
| 2nvk.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.07 | Thioredoxin Reductase |
| 1fl2.1.A | 16.13 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | ALKYL HYDROPEROXIDE REDUCTASE SUBUNIT F |
| 2rab.1.A | 15.63 | homo-dimer | HHblits | X-ray | 2.50Å | 0.25 | 0.07 | glutathione amide reductase |
| 1lrj.1.A | 22.58 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.07 | UDP-glucose 4-epimerase |
| 1orr.1.A | 22.58 | homo-tetramer | HHblits | X-ray | 1.50Å | 0.27 | 0.07 | CDP-tyvelose-2-epimerase |
| 4a5l.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.66Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 4a5l.1.B | 12.50 | homo-dimer | HHblits | X-ray | 1.66Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 4ccr.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.28Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 4up3.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.44Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 4up3.1.B | 12.50 | homo-dimer | HHblits | X-ray | 1.44Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 3uie.1.A | 26.67 | homo-dimer | HHblits | X-ray | 1.79Å | 0.30 | 0.07 | Adenylyl-sulfate kinase 1, chloroplastic |
| 2c9y.1.A | 19.35 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.07 | ADENYLATE KINASE ISOENZYME 2, MITOCHONDRIAL |
| 3bsy.1.A | 16.13 | homo-trimer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | Acetyltransferase |
| 3bsw.1.A | 16.13 | homo-trimer | HHblits | X-ray | 1.77Å | 0.27 | 0.07 | Acetyltransferase |
| 3bss.1.A | 16.13 | homo-trimer | HHblits | X-ray | 2.30Å | 0.27 | 0.07 | Acetyltransferase |
| 3bsy.1.B | 16.13 | homo-trimer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | Acetyltransferase |
| 2vhe.1.A | 16.13 | homo-trimer | HHblits | X-ray | 1.80Å | 0.27 | 0.07 | ACETYLTRANSFERASE |
| 3bfp.1.A | 16.13 | homo-trimer | HHblits | X-ray | 1.75Å | 0.27 | 0.07 | Acetyltransferase |
| 5glg.1.A | 15.63 | monomer | HHblits | X-ray | 1.80Å | 0.24 | 0.07 | Fumarate reductase 2 |
| 4jnq.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.80Å | 0.24 | 0.07 | Thioredoxin reductase |
| 1tdf.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.30Å | 0.24 | 0.07 | THIOREDOXIN REDUCTASE |
| 1cl0.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.07 | THIOREDOXIN REDUCTASE |
| 1trb.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.07 | THIOREDOXIN REDUCTASE |
| 2vli.1.A | 23.33 | homo-dimer | HHblits | X-ray | 1.95Å | 0.29 | 0.07 | ANTIBIOTIC RESISTANCE PROTEIN |
| 2vli.2.A | 23.33 | homo-dimer | HHblits | X-ray | 1.95Å | 0.29 | 0.07 | ANTIBIOTIC RESISTANCE PROTEIN |
| 4k46.1.A | 16.67 | monomer | HHblits | X-ray | 2.01Å | 0.29 | 0.07 | Adenylate kinase |
| 3f8d.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.40Å | 0.24 | 0.07 | Thioredoxin reductase (TrxB-3) |
| 3f8p.1.A | 15.63 | homo-dimer | HHblits | X-ray | 1.80Å | 0.24 | 0.07 | Thioredoxin reductase (TrxB-3) |
| 3f8r.1.B | 15.63 | homo-dimer | HHblits | X-ray | 1.95Å | 0.24 | 0.07 | Thioredoxin reductase (TrxB-3) |
| 5ncc.1.A | 18.75 | monomer | HHblits | X-ray | 3.12Å | 0.24 | 0.07 | Fatty acid Photodecarboxylase |
| 4loa.1.A | 39.29 | homo-dimer | HHblits | X-ray | 1.82Å | 0.35 | 0.06 | De-novo design amidase |
| 4loa.1.B | 39.29 | homo-dimer | HHblits | X-ray | 1.82Å | 0.35 | 0.06 | De-novo design amidase |
| 4ttq.1.A | 31.03 | monomer | HHblits | X-ray | 2.20Å | 0.32 | 0.07 | Dephospho-CoA kinase |
| 4ttp.1.A | 31.03 | monomer | HHblits | X-ray | 2.20Å | 0.32 | 0.07 | Dephospho-CoA kinase |
| 3ean.1.A | 16.13 | homo-dimer | HHblits | X-ray | 2.75Å | 0.26 | 0.07 | Thioredoxin reductase 1 |
| 1h6v.1.A | 16.13 | homo-dimer | HHblits | X-ray | 3.00Å | 0.26 | 0.07 | THIOREDOXIN REDUCTASE |
| 4kpr.1.A | 16.13 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | Thioredoxin reductase 1, cytoplasmic |
| 2whd.1.A | 9.38 | homo-dimer | HHblits | X-ray | 2.60Å | 0.24 | 0.07 | THIOREDOXIN REDUCTASE |
| 3lzw.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.80Å | 0.24 | 0.07 | Ferredoxin--NADP reductase 2 |
| 3lzx.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.90Å | 0.24 | 0.07 | Ferredoxin--NADP reductase 2 |
| 1zmx.1.A | 12.50 | homo-dimer | HHblits | X-ray | 3.10Å | 0.24 | 0.07 | Deoxynucleoside kinase |
| 1zm7.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.07 | Deoxynucleoside kinase |
| 3v3t.1.A | 15.63 | monomer | HHblits | X-ray | 2.30Å | 0.24 | 0.07 | Cell division GTPase FtsZ, diverged |
| 1e2e.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.07 | THYMIDYLATE KINASE |
| 1e9c.1.A | 20.00 | homo-dimer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | THYMIDYLATE KINASE |
| 1e4v.1.A | 16.67 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.07 | ADENYLATE KINASE |
| 3kjg.2.A | 20.69 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kjg.1.A | 20.69 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kji.1.A | 20.69 | homo-dimer | HHblits | X-ray | 2.13Å | 0.31 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kji.2.A | 20.69 | homo-dimer | HHblits | X-ray | 2.13Å | 0.31 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kjh.1.A | 20.69 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 3kje.1.A | 20.69 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC |
| 2cdu.1.A | 12.90 | homo-dimer | HHblits | X-ray | 1.80Å | 0.26 | 0.07 | NADPH OXIDASE |
| 3fb4.1.A | 16.67 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | Adenylate kinase |
| 2xx3.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.07 | THYMIDYLATE KINASE |
| 4ag6.1.A | 23.33 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.07 | TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN |
| 4ag6.1.B | 23.33 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.07 | TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN |
| 4ag6.2.B | 23.33 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.07 | TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN |
| 4ag5.2.B | 23.33 | homo-dimer | HHblits | X-ray | 2.45Å | 0.28 | 0.07 | TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN |
| 2v3a.1.A | 9.38 | monomer | HHblits | X-ray | 2.40Å | 0.23 | 0.07 | RUBREDOXIN REDUCTASE |
| 2v3b.1.A | 9.38 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.23 | 0.07 | RUBREDOXIN REDUCTASE |
| 4pfs.1.A | 24.14 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | Cobyrinic Acid a,c-diamide synthase |
| 4pfs.2.A | 24.14 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | Cobyrinic Acid a,c-diamide synthase |
| 5if9.1.A | 24.14 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.07 | Cobyrinic Acid a,c-diamide synthase |
| 4cvn.1.A | 28.57 | hetero-oligomer | HHblits | X-ray | 2.12Å | 0.33 | 0.06 | PUTATIVE ADENYLATE KINASE |
| 4zrm.1.A | 22.58 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 4zrn.1.A | 22.58 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 4new.1.A | 16.13 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.07 | Trypanothione reductase, putative |
| 4bzp.1.A | 34.62 | homo-dimer | HHblits | X-ray | 1.47Å | 0.39 | 0.06 | BIFUNCTIONAL ENZYME CYSN/CYSC |
| 4bzq.1.A | 34.62 | homo-dimer | HHblits | X-ray | 2.10Å | 0.39 | 0.06 | BIFUNCTIONAL ENZYME CYSN/CYSC |
| 1kvs.1.A | 19.35 | monomer | HHblits | X-ray | 2.15Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1kvt.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1kvr.1.A | 19.35 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1lrl.1.A | 19.35 | homo-dimer | HHblits | X-ray | 1.80Å | 0.25 | 0.07 | UDP-glucose 4-epimerase |
| 1a9z.1.A | 19.35 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1f8w.1.A | 12.90 | homo-tetramer | HHblits | X-ray | 2.45Å | 0.25 | 0.07 | NADH PEROXIDASE |
| 1joa.1.A | 12.90 | monomer | HHblits | X-ray | 2.80Å | 0.25 | 0.07 | NADH PEROXIDASE |
| 1nhp.1.A | 12.90 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.07 | NADH PEROXIDASE |
| 1nhq.1.A | 12.90 | monomer | HHblits | X-ray | 2.00Å | 0.25 | 0.07 | NADH PEROXIDASE |
| 3n2e.1.A | 39.29 | monomer | HHblits | X-ray | 2.53Å | 0.33 | 0.06 | Shikimate kinase |
| 2rir.1.A | 12.90 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.25 | 0.07 | Dipicolinate synthase, A chain |
| 2rir.1.B | 12.90 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.25 | 0.07 | Dipicolinate synthase, A chain |
| 4qbi.1.A | 20.69 | monomer | HHblits | X-ray | 1.67Å | 0.30 | 0.07 | Adenylate kinase |
| 4qbi.2.A | 20.69 | monomer | HHblits | X-ray | 1.67Å | 0.30 | 0.07 | Adenylate kinase |
| 5x3x.1.A | 24.14 | hetero-oligomer | HHblits | X-ray | 2.79Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x3x.1.B | 24.14 | hetero-oligomer | HHblits | X-ray | 2.79Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x3x.2.A | 24.14 | hetero-oligomer | HHblits | X-ray | 2.79Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x3x.2.B | 24.14 | hetero-oligomer | HHblits | X-ray | 2.79Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x41.1.A | 24.14 | hetero-oligomer | HHblits | X-ray | 3.47Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x41.1.B | 24.14 | hetero-oligomer | HHblits | X-ray | 3.47Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x41.2.A | 24.14 | hetero-oligomer | HHblits | X-ray | 3.47Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 5x41.2.B | 24.14 | hetero-oligomer | HHblits | X-ray | 3.47Å | 0.30 | 0.07 | Cobalt ABC transporter ATP-binding protein |
| 1kvq.1.A | 19.35 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1a9y.1.A | 19.35 | homo-dimer | HHblits | X-ray | 1.80Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 1udc.1.A | 19.35 | homo-dimer | HHblits | X-ray | 1.65Å | 0.25 | 0.07 | UDP-GALACTOSE-4-EPIMERASE |
| 1kvu.1.A | 19.35 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.07 | UDP-GALACTOSE 4-EPIMERASE |
| 3d8x.1.A | 9.68 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.07 | Thioredoxin reductase 1 |
| 3itj.1.A | 9.68 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.07 | Thioredoxin reductase 1 |
| 3itj.1.B | 9.68 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.07 | Thioredoxin reductase 1 |
| 3itj.2.B | 9.68 | homo-dimer | HHblits | X-ray | 2.40Å | 0.25 | 0.07 | Thioredoxin reductase 1 |
| 1f6m.1.A | 12.90 | hetero-oligomer | HHblits | X-ray | 2.95Å | 0.25 | 0.07 | THIOREDOXIN REDUCTASE |
| 3uxm.1.A | 16.67 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.07 | Thymidylate kinase |
| 3uxm.2.A | 16.67 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.07 | Thymidylate kinase |
| 1e6c.1.A | 25.00 | monomer | HHblits | X-ray | 1.80Å | 0.32 | 0.06 | SHIKIMATE KINASE |
| 2wba.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2wpc.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2wpc.2.A | 16.67 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2wp5.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 1dvr.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.36Å | 0.27 | 0.07 | ADENYLATE KINASE |
| 1dvr.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.36Å | 0.27 | 0.07 | ADENYLATE KINASE |
| 3vaa.1.A | 13.33 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.07 | Shikimate kinase |
| 3vaa.2.A | 13.33 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.07 | Shikimate kinase |
| 3vaa.3.A | 13.33 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.07 | Shikimate kinase |
| 1zak.1.A | 12.90 | homo-dimer | HHblits | X-ray | 3.50Å | 0.24 | 0.07 | ADENYLATE KINASE |
| 3dl0.1.A | 17.24 | monomer | HHblits | X-ray | 1.58Å | 0.29 | 0.07 | Adenylate kinase |
| 3lad.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 3lad.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.07 | DIHYDROLIPOAMIDE DEHYDROGENASE |
| 1vdc.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.07 | NADPH DEPENDENT THIOREDOXIN REDUCTASE |
| 1p3j.1.A | 17.24 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | Adenylate kinase |
| 2oo7.1.A | 17.24 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Adenylate kinase |
| 5g3y.1.A | 20.69 | monomer | HHblits | X-ray | 1.18Å | 0.29 | 0.07 | ADENYLATE KINSE |
| 2a30.1.A | 20.69 | homo-dimer | HHblits | X-ray | 3.02Å | 0.29 | 0.07 | Deoxycytidine kinase |
| 2ak3.1.A | 17.24 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.07 | ADENYLATE KINASE ISOENZYME-3 |
| 2ak3.2.A | 17.24 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.07 | ADENYLATE KINASE ISOENZYME-3 |
| 1knx.1.A | 17.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.B | 17.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.C | 17.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.D | 17.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.E | 17.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1knx.1.F | 17.24 | homo-hexamer | HHblits | X-ray | 2.50Å | 0.29 | 0.07 | Probable HPr(Ser) kinase/phosphatase |
| 1ot3.2.A | 12.90 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.07 | Deoxyribonucleoside Kinase |
| 2vp0.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.20Å | 0.24 | 0.07 | DEOXYNUCLEOSIDE KINASE |
| 1ot3.2.B | 12.90 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.07 | Deoxyribonucleoside Kinase |
| 2jcs.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.50Å | 0.24 | 0.07 | DEOXYNUCLEOSIDE KINASE |
| 2eu8.1.A | 17.24 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Adenylate kinase |
| 2eu8.2.A | 17.24 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.07 | Adenylate kinase |
| 3tau.1.A | 17.24 | homo-dimer | HHblits | X-ray | 2.05Å | 0.29 | 0.07 | Guanylate kinase |
| 3tau.1.B | 17.24 | homo-dimer | HHblits | X-ray | 2.05Å | 0.29 | 0.07 | Guanylate kinase |
| 1zd8.1.A | 17.24 | monomer | HHblits | X-ray | 1.48Å | 0.29 | 0.07 | GTP:AMP phosphotransferase mitochondrial |
| 4mkh.1.A | 25.00 | monomer | HHblits | X-ray | 1.50Å | 0.31 | 0.06 | Adenylate kinase |
| 1gxf.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) |
| 1bzl.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) |
| 1aog.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 1nda.1.A | 16.67 | homo-dimer | HHblits | X-ray | 3.30Å | 0.26 | 0.07 | TRYPANOTHIONE OXIDOREDUCTASE |
| 1e98.1.A | 20.69 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.07 | THYMIDYLATE KINASE |
| 1nmy.1.A | 20.69 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.07 | similar to THYMIDYLATE KINASE (DTMP KINASE) |
| 1p9r.1.A | 28.57 | monomer | HHblits | X-ray | 2.50Å | 0.31 | 0.06 | General secretion pathway protein E |
| 1typ.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2tpr.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2tpr.1.B | 13.33 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 1fea.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 1fea.1.B | 13.33 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 1tyt.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.07 | TRYPANOTHIONE REDUCTASE, OXIDIZED FORM |
| 2qmo.1.A | 13.33 | homo-dimer | HHblits | X-ray | 1.47Å | 0.26 | 0.07 | Dethiobiotin synthetase |
| 2axp.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.07 | hypothetical protein BSU20280 |
| 3kb2.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | SPBc2 prophage-derived uncharacterized protein yorR |
| 3kb2.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.07 | SPBc2 prophage-derived uncharacterized protein yorR |
| 5xb5.1.A | 27.59 | homo-dimer | HHblits | X-ray | 2.23Å | 0.28 | 0.07 | Thymidylate kinase |
| 2pbr.1.B | 27.59 | homo-dimer | HHblits | X-ray | 1.96Å | 0.28 | 0.07 | Thymidylate kinase |
| 2pbr.1.A | 27.59 | homo-dimer | HHblits | X-ray | 1.96Å | 0.28 | 0.07 | Thymidylate kinase |
| 4s35.1.B | 27.59 | homo-dimer | HHblits | X-ray | 1.55Å | 0.28 | 0.07 | Thymidylate kinase |
| 5xb2.1.B | 27.59 | homo-dimer | HHblits | X-ray | 2.16Å | 0.28 | 0.07 | Thymidylate kinase |
| 4y0a.1.A | 25.00 | monomer | HHblits | X-ray | 1.91Å | 0.31 | 0.06 | Shikimate kinase |
| 2x50.1.A | 13.33 | homo-dimer | HHblits | X-ray | 3.30Å | 0.25 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2yau.1.A | 13.33 | homo-dimer | HHblits | X-ray | 3.50Å | 0.25 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 2jk6.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.95Å | 0.25 | 0.07 | TRYPANOTHIONE REDUCTASE |
| 3c8u.1.A | 25.93 | monomer | HHblits | X-ray | 1.95Å | 0.33 | 0.06 | Fructokinase |
| 3dkv.1.A | 21.43 | monomer | HHblits | X-ray | 1.82Å | 0.30 | 0.06 | Adenylate kinase |
| 1a2b.1.A | 25.00 | monomer | HHblits | X-ray | 2.40Å | 0.30 | 0.06 | TRANSFORMING PROTEIN RHOA |
| 1cxz.1.A | 25.00 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.30 | 0.06 | PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)) |
| 4qbf.1.A | 21.43 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.06 | Adenylate kinase |
| 2eyu.1.A | 25.93 | monomer | HHblits | X-ray | 1.87Å | 0.33 | 0.06 | twitching motility protein PilT |
| 2eyu.2.A | 25.93 | monomer | HHblits | X-ray | 1.87Å | 0.33 | 0.06 | twitching motility protein PilT |
| 4gmd.1.B | 17.24 | homo-dimer | HHblits | X-ray | 1.98Å | 0.27 | 0.07 | Thymidylate kinase |
| 4gmd.1.A | 17.24 | homo-dimer | HHblits | X-ray | 1.98Å | 0.27 | 0.07 | Thymidylate kinase |
| 4edh.1.A | 17.24 | homo-dimer | HHblits | X-ray | 1.32Å | 0.27 | 0.07 | Thymidylate kinase |
| 4e5u.1.A | 17.24 | homo-dimer | HHblits | X-ray | 1.81Å | 0.27 | 0.07 | Thymidylate kinase |
| 4e5u.1.B | 17.24 | homo-dimer | HHblits | X-ray | 1.81Å | 0.27 | 0.07 | Thymidylate kinase |
| 2yvu.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.06 | Probable adenylyl-sulfate kinase |
| 2yvu.1.B | 25.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.06 | Probable adenylyl-sulfate kinase |
| 2vp9.1.A | 13.33 | homo-dimer | HHblits | X-ray | 2.90Å | 0.24 | 0.07 | DEOXYNUCLEOSIDE KINASE |
| 2vp4.1.A | 13.33 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.24 | 0.07 | DEOXYNUCLEOSIDE KINASE |
| 2vp5.1.B | 13.33 | homo-dimer | HHblits | X-ray | 2.30Å | 0.24 | 0.07 | DEOXYNUCLEOSIDE KINASE |
| 3msx.1.A | 25.00 | hetero-oligomer | HHblits | X-ray | 1.65Å | 0.30 | 0.06 | Transforming protein RhoA |
| 1ki6.1.A | 40.00 | hetero-oligomer | HHblits | X-ray | 2.37Å | 0.38 | 0.06 | THYMIDINE KINASE |
| 1a7j.1.A | 22.22 | monomer | HHblits | X-ray | 2.50Å | 0.32 | 0.06 | PHOSPHORIBULOKINASE |
| 2p3s.1.A | 17.86 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.06 | Adenylate kinase |
| 2qaj.1.A | 17.86 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.06 | Adenylate kinase |
| 2ori.1.A | 17.86 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.06 | Adenylate kinase |
| 2ori.1.B | 17.86 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.06 | Adenylate kinase |
| 3hjn.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.06 | Thymidylate kinase |
| 3hjn.1.B | 25.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.06 | Thymidylate kinase |
| 3h86.2.A | 29.63 | homo-trimer | HHblits | X-ray | 2.50Å | 0.32 | 0.06 | Adenylate kinase |
| 3h86.1.B | 29.63 | homo-trimer | HHblits | X-ray | 2.50Å | 0.32 | 0.06 | Adenylate kinase |
| 3h86.1.C | 29.63 | homo-trimer | HHblits | X-ray | 2.50Å | 0.32 | 0.06 | Adenylate kinase |
| 3h86.1.A | 29.63 | homo-trimer | HHblits | X-ray | 2.50Å | 0.32 | 0.06 | Adenylate kinase |
| 1zip.1.A | 17.86 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.06 | ADENYLATE KINASE |
| 2go1.1.A | 21.43 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.06 | NAD-dependent formate dehydrogenase |
| 2gug.1.A | 21.43 | homo-dimer | HHblits | X-ray | 2.28Å | 0.29 | 0.06 | Formate dehydrogenase |
| 4ypn.1.A | 18.52 | monomer | HHblits | X-ray | 2.07Å | 0.31 | 0.06 | Lon protease |
| 3igf.1.A | 13.79 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | All4481 protein |
| 3igf.1.B | 13.79 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.07 | All4481 protein |
| 3tqc.1.A | 34.62 | homo-dimer | HHblits | X-ray | 2.30Å | 0.34 | 0.06 | Pantothenate kinase |
| 2plr.1.A | 44.00 | homo-dimer | HHblits | X-ray | 1.60Å | 0.37 | 0.06 | Probable thymidylate kinase |
| 2plr.1.B | 44.00 | homo-dimer | HHblits | X-ray | 1.60Å | 0.37 | 0.06 | Probable thymidylate kinase |
| 4rzu.1.A | 44.00 | homo-dimer | HHblits | X-ray | 2.80Å | 0.37 | 0.06 | Probable thymidylate kinase |
| 4rzu.1.B | 44.00 | homo-dimer | HHblits | X-ray | 2.80Å | 0.37 | 0.06 | Probable thymidylate kinase |
| 4rzx.1.B | 44.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.37 | 0.06 | Probable thymidylate kinase |
| 3wr5.1.A | 21.43 | homo-dimer | HHblits | X-ray | 2.14Å | 0.28 | 0.06 | Formate dehydrogenase |
| 3wr5.2.B | 21.43 | homo-dimer | HHblits | X-ray | 2.14Å | 0.28 | 0.06 | Formate dehydrogenase |
| 2rhm.1.A | 21.43 | homo-dimer | HHblits | X-ray | 1.70Å | 0.28 | 0.06 | Putative kinase |
| 3d4o.1.A | 10.34 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.07 | Dipicolinate synthase subunit A |
| 3d4o.1.B | 10.34 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.07 | Dipicolinate synthase subunit A |
| 5g3z.1.A | 22.22 | monomer | HHblits | X-ray | 1.89Å | 0.31 | 0.06 | ADENYLATE KINSE |
| 5g40.1.A | 22.22 | monomer | HHblits | X-ray | 1.69Å | 0.31 | 0.06 | ADENYLATE KINSE |
| 5g41.1.A | 22.22 | monomer | HHblits | X-ray | 1.54Å | 0.31 | 0.06 | ADENYLATE KINSE |
| 2v54.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.40Å | 0.30 | 0.06 | THYMIDYLATE KINASE |
| 1dpf.1.A | 25.93 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.06 | RHOA |
| 2le0.1.A | 14.81 | monomer | HHblits | NMR | NA | 0.30 | 0.06 | Poly [ADP-ribose] polymerase 1 |
| 2i3b.1.A | 30.77 | monomer | HHblits | NMR | NA | 0.33 | 0.06 | Human Cancer-Related NTPase |
| 4bzq.1.A | 25.93 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.06 | BIFUNCTIONAL ENZYME CYSN/CYSC |
| 1vpl.1.A | 23.08 | homo-dimer | HHblits | X-ray | 2.10Å | 0.33 | 0.06 | ABC transporter, ATP-binding protein |
| 3fdi.1.A | 21.43 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.06 | uncharacterized protein |
| 1s3g.1.A | 22.22 | monomer | HHblits | X-ray | 2.25Å | 0.29 | 0.06 | Adenylate kinase |
| 1e9f.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.06 | THYMIDYLATE KINASE |
| 5xbh.1.A | 29.63 | homo-dimer | HHblits | X-ray | 2.23Å | 0.29 | 0.06 | Thymidylate kinase |
| 4f7w.1.B | 36.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.06 | Pantothenate kinase |
| 4f7w.1.A | 36.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.06 | Pantothenate kinase |
| 4f7w.3.A | 36.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.06 | Pantothenate kinase |
| 4f7w.4.B | 36.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.06 | Pantothenate kinase |
| 4ne2.2.A | 36.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.35 | 0.06 | Pantothenate kinase |
| 1sq5.1.A | 36.00 | homo-dimer | HHblits | X-ray | 2.20Å | 0.35 | 0.06 | Pantothenate kinase |
| 1esm.1.A | 36.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.35 | 0.06 | PANTOTHENATE KINASE |
| 1esm.1.B | 36.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.35 | 0.06 | PANTOTHENATE KINASE |
| 1esn.1.A | 36.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.35 | 0.06 | PANTOTHENATE KINASE |
| 1esn.1.B | 36.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.35 | 0.06 | PANTOTHENATE KINASE |
| 1esn.2.B | 36.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.35 | 0.06 | PANTOTHENATE KINASE |
| 4typ.1.A | 18.52 | monomer | HHblits | X-ray | 2.90Å | 0.29 | 0.06 | Adenylate kinase |
| 5tmp.1.A | 23.08 | monomer | HHblits | X-ray | 1.98Å | 0.32 | 0.06 | PROTEIN (THYMIDYLATE KINASE) |
| 3ld9.1.A | 36.00 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.35 | 0.06 | Thymidylate kinase |
| 3avo.1.A | 30.77 | homo-dimer | HHblits | X-ray | 2.55Å | 0.32 | 0.06 | Pantothenate kinase |
| 5tiz.1.A | 14.29 | monomer | HHblits | X-ray | 2.87Å | 0.26 | 0.06 | Sulfotransferase |
| 3jvv.1.A | 28.00 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.34 | 0.06 | Twitching mobility protein |
| 3jvv.1.B | 28.00 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.34 | 0.06 | Twitching mobility protein |
| 3jvv.1.C | 28.00 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.34 | 0.06 | Twitching mobility protein |
| 3jvu.1.A | 28.00 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.34 | 0.06 | Twitching mobility protein |
| 3jvu.1.B | 28.00 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.34 | 0.06 | Twitching mobility protein |
| 3jvu.1.C | 28.00 | homo-hexamer | HHblits | X-ray | 3.10Å | 0.34 | 0.06 | Twitching mobility protein |
| 4rfv.1.A | 39.13 | homo-dimer | HHblits | X-ray | 1.69Å | 0.41 | 0.05 | Bifunctional enzyme CysN/CysC |
| 5x40.1.A | 26.92 | homo-dimer | HHblits | X-ray | 1.45Å | 0.31 | 0.06 | Cobalt ABC transporter ATP-binding protein |
| 5x40.1.B | 26.92 | homo-dimer | HHblits | X-ray | 1.45Å | 0.31 | 0.06 | Cobalt ABC transporter ATP-binding protein |
| 3uwo.1.A | 18.52 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.06 | Thymidylate kinase |
| 1ki9.1.A | 30.77 | homo-trimer | HHblits | X-ray | 2.76Å | 0.31 | 0.06 | adenylate kinase |
| 1ki9.1.B | 30.77 | homo-trimer | HHblits | X-ray | 2.76Å | 0.31 | 0.06 | adenylate kinase |
| 1ki9.1.C | 30.77 | homo-trimer | HHblits | X-ray | 2.76Å | 0.31 | 0.06 | adenylate kinase |
| 2yof.1.A | 22.22 | monomer | HHblits | X-ray | 1.82Å | 0.28 | 0.06 | THYMIDYLATE KINASE |
| 2yof.3.A | 22.22 | monomer | HHblits | X-ray | 1.82Å | 0.28 | 0.06 | THYMIDYLATE KINASE |
| 2yog.1.A | 22.22 | monomer | HHblits | X-ray | 1.50Å | 0.28 | 0.06 | THYMIDYLATE KINASE |
| 2yog.2.A | 22.22 | monomer | HHblits | X-ray | 1.50Å | 0.28 | 0.06 | THYMIDYLATE KINASE |
| 2wwg.2.B | 22.22 | homo-dimer | HHblits | X-ray | 2.40Å | 0.28 | 0.06 | THYMIDILATE KINASE, PUTATIVE |
| 2wwg.2.A | 22.22 | homo-dimer | HHblits | X-ray | 2.40Å | 0.28 | 0.06 | THYMIDILATE KINASE, PUTATIVE |
| 5b3f.1.A | 28.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.34 | 0.06 | Phosphoribulokinase/uridine kinase |
| 5b3f.1.B | 28.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.34 | 0.06 | Phosphoribulokinase/uridine kinase |
| 4bzp.1.A | 26.92 | homo-dimer | HHblits | X-ray | 1.47Å | 0.31 | 0.06 | BIFUNCTIONAL ENZYME CYSN/CYSC |
| 4gfd.1.A | 19.23 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.06 | Thymidylate kinase |
| 2cck.1.B | 19.23 | homo-dimer | HHblits | X-ray | 2.21Å | 0.31 | 0.06 | THYMIDYLATE KINASE |
| 2plr.1.A | 30.77 | homo-dimer | HHblits | X-ray | 1.60Å | 0.31 | 0.06 | Probable thymidylate kinase |
| 2plr.1.B | 30.77 | homo-dimer | HHblits | X-ray | 1.60Å | 0.31 | 0.06 | Probable thymidylate kinase |
| 4rzu.1.A | 30.77 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.06 | Probable thymidylate kinase |
| 4rzu.1.B | 30.77 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.06 | Probable thymidylate kinase |
| 4rzx.1.B | 30.77 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.06 | Probable thymidylate kinase |
| 1kht.1.A | 26.92 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.06 | adenylate kinase |
| 1kht.1.B | 26.92 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.06 | adenylate kinase |
| 1kht.1.C | 26.92 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.06 | adenylate kinase |
| 1grr.1.A | 14.81 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.27 | 0.06 | CHLORAMPHENICOL 3-O PHOSPHOTRANSFERASE |
| 1grq.1.A | 14.81 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.27 | 0.06 | CHLORAMPHENICOL 3-O PHOSPHOTRANSFERASE |
| 2vli.1.A | 36.00 | homo-dimer | HHblits | X-ray | 1.95Å | 0.33 | 0.06 | ANTIBIOTIC RESISTANCE PROTEIN |
| 2vli.2.A | 36.00 | homo-dimer | HHblits | X-ray | 1.95Å | 0.33 | 0.06 | ANTIBIOTIC RESISTANCE PROTEIN |
| 1tmk.1.A | 19.23 | homo-dimer | HHblits | X-ray | 2.10Å | 0.30 | 0.06 | THYMIDYLATE KINASE |
| 3tmk.1.A | 19.23 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.06 | THYMIDYLATE KINASE |
| 3tmk.1.B | 19.23 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.06 | THYMIDYLATE KINASE |
| 5uiv.1.A | 28.00 | homo-dimer | HHblits | X-ray | 2.45Å | 0.33 | 0.06 | Bifunctional thymidylate/uridylate kinase |
| 1xrj.1.A | 28.00 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.33 | 0.06 | Uridine-cytidine kinase 2 |
| 1ufq.1.A | 28.00 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.33 | 0.06 | Uridine-cytidine kinase 2 |
| 1uej.1.A | 28.00 | monomer | HHblits | X-ray | 2.61Å | 0.33 | 0.06 | Uridine-cytidine kinase 2 |
| 1ufq.1.B | 28.00 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.33 | 0.06 | Uridine-cytidine kinase 2 |
| 1uj2.1.A | 28.00 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.33 | 0.06 | Uridine-cytidine kinase 2 |
| 1lvg.1.A | 19.23 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.06 | Guanylate kinase |
| 1e9f.1.A | 28.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.32 | 0.06 | THYMIDYLATE KINASE |
| 5cb6.3.B | 16.00 | homo-dimer | HHblits | X-ray | 2.79Å | 0.32 | 0.06 | Probable adenylyl-sulfate kinase |
| 5cb6.2.B | 16.00 | homo-dimer | HHblits | X-ray | 2.79Å | 0.32 | 0.06 | Probable adenylyl-sulfate kinase |
| 1q3t.1.A | 20.00 | monomer | HHblits | NMR | NA | 0.32 | 0.06 | Cytidylate kinase |
| 3tr0.1.A | 24.00 | homo-dimer | HHblits | X-ray | 1.85Å | 0.32 | 0.06 | Guanylate kinase |
| 1odf.1.A | 23.08 | monomer | HHblits | X-ray | 2.25Å | 0.28 | 0.06 | HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION |
| 3sqk.1.A | 19.23 | monomer | HHblits | X-ray | 2.45Å | 0.28 | 0.06 | Guanylate kinase |
| 3sqk.2.A | 19.23 | monomer | HHblits | X-ray | 2.45Å | 0.28 | 0.06 | Guanylate kinase |
| 4rfv.1.A | 28.00 | homo-dimer | HHblits | X-ray | 1.69Å | 0.31 | 0.06 | Bifunctional enzyme CysN/CysC |
| 4cvn.1.A | 43.48 | hetero-oligomer | HHblits | X-ray | 2.12Å | 0.38 | 0.05 | PUTATIVE ADENYLATE KINASE |
| 3lnc.1.A | 15.38 | homo-dimer | HHblits | X-ray | 1.95Å | 0.28 | 0.06 | Guanylate kinase |
| 4mqb.1.A | 28.00 | homo-dimer | HHblits | X-ray | 1.55Å | 0.31 | 0.06 | Thymidylate kinase |
| 4mqb.1.B | 28.00 | homo-dimer | HHblits | X-ray | 1.55Å | 0.31 | 0.06 | Thymidylate kinase |
| 4gfd.1.A | 28.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.06 | Thymidylate kinase |
| 2cck.1.B | 28.00 | homo-dimer | HHblits | X-ray | 2.21Å | 0.31 | 0.06 | THYMIDYLATE KINASE |
| 1tmk.1.A | 28.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.06 | THYMIDYLATE KINASE |
| 3tmk.1.A | 28.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.06 | THYMIDYLATE KINASE |
| 3tmk.1.B | 28.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.06 | THYMIDYLATE KINASE |
| 4y0a.1.A | 29.17 | monomer | HHblits | X-ray | 1.91Å | 0.34 | 0.05 | Shikimate kinase |
| 2qor.1.A | 15.38 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.06 | Guanylate kinase |
| 3dhw.1.C | 15.38 | hetero-oligomer | HHblits | X-ray | 3.70Å | 0.28 | 0.06 | Methionine import ATP-binding protein metN |
| 3dhw.1.D | 15.38 | hetero-oligomer | HHblits | X-ray | 3.70Å | 0.28 | 0.06 | Methionine import ATP-binding protein metN |
| 3dhw.2.C | 15.38 | hetero-oligomer | HHblits | X-ray | 3.70Å | 0.28 | 0.06 | Methionine import ATP-binding protein metN |
| 3dhw.2.D | 15.38 | hetero-oligomer | HHblits | X-ray | 3.70Å | 0.28 | 0.06 | Methionine import ATP-binding protein metN |
| 3ipy.1.A | 20.83 | monomer | HHblits | X-ray | 2.54Å | 0.34 | 0.05 | Deoxycytidine kinase |
| 3ipx.1.A | 20.83 | monomer | HHblits | X-ray | 2.00Å | 0.34 | 0.05 | Deoxycytidine kinase |
| 3cr7.1.A | 29.17 | homo-dimer | HHblits | X-ray | 2.50Å | 0.34 | 0.05 | Adenylyl-sulfate kinase |
| 3cr7.1.B | 29.17 | homo-dimer | HHblits | X-ray | 2.50Å | 0.34 | 0.05 | Adenylyl-sulfate kinase |
| 3r20.1.A | 33.33 | monomer | HHblits | X-ray | 2.00Å | 0.34 | 0.05 | Cytidylate kinase |
| 1y63.1.A | 34.78 | monomer | HHblits | X-ray | 1.70Å | 0.37 | 0.05 | Lmaj004144AAA protein |
| 3asz.1.A | 29.17 | homo-dimer | HHblits | X-ray | 2.25Å | 0.33 | 0.05 | Uridine kinase |
| 3asy.1.B | 29.17 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.05 | Uridine kinase |
| 3asy.1.A | 29.17 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.05 | Uridine kinase |
| 3asz.1.B | 29.17 | homo-dimer | HHblits | X-ray | 2.25Å | 0.33 | 0.05 | Uridine kinase |
| 3v9p.1.A | 23.08 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.06 | Thymidylate kinase |
| 1kht.1.A | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.33 | 0.05 | adenylate kinase |
| 1kht.1.B | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.33 | 0.05 | adenylate kinase |
| 1kht.1.C | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.33 | 0.05 | adenylate kinase |
| 5tmp.1.A | 25.00 | monomer | HHblits | X-ray | 1.98Å | 0.33 | 0.05 | PROTEIN (THYMIDYLATE KINASE) |
| 2ak2.1.A | 20.00 | monomer | HHblits | X-ray | 2.10Å | 0.30 | 0.06 | ADENYLATE KINASE ISOENZYME-2 |
| 2f1r.1.A | 20.83 | homo-dimer | HHblits | X-ray | 2.10Å | 0.32 | 0.05 | molybdopterin-guanine dinucleotide biosynthesis protein B (mobB) |
| 2f1r.1.B | 20.83 | homo-dimer | HHblits | X-ray | 2.10Å | 0.32 | 0.05 | molybdopterin-guanine dinucleotide biosynthesis protein B (mobB) |
| 2v54.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.06 | THYMIDYLATE KINASE |
| 1s96.1.A | 24.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.06 | Guanylate kinase |
| 1ex7.1.A | 20.00 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.06 | GUANYLATE KINASE |
| 1ex6.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.06 | GUANYLATE KINASE |
| 1ex6.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.06 | GUANYLATE KINASE |
| 1kdo.1.A | 20.83 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 1kdo.2.A | 20.83 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 1kdt.1.A | 20.83 | monomer | HHblits | X-ray | 1.95Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 1kdt.2.A | 20.83 | monomer | HHblits | X-ray | 1.95Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 1kdp.2.A | 20.83 | monomer | HHblits | X-ray | 2.30Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 1kdr.1.A | 20.83 | monomer | HHblits | X-ray | 2.25Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 1kdr.2.A | 20.83 | monomer | HHblits | X-ray | 2.25Å | 0.32 | 0.05 | CYTIDYLATE KINASE |
| 2cmk.1.A | 20.83 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.05 | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE) |
| 2fem.1.A | 20.83 | monomer | HHblits | X-ray | 1.90Å | 0.32 | 0.05 | Cytidylate kinase |
| 2feo.1.A | 20.83 | monomer | HHblits | X-ray | 2.80Å | 0.32 | 0.05 | Cytidylate kinase |
| 1cke.1.A | 20.83 | monomer | HHblits | X-ray | 1.75Å | 0.32 | 0.05 | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE) |
| 1rkb.1.A | 29.17 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.05 | Protein AD-004 |
| 3t61.1.A | 29.17 | homo-dimer | HHblits | X-ray | 2.20Å | 0.32 | 0.05 | Gluconokinase |
| 3w8r.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.32 | 0.05 | Uridine kinase |
| 2qsy.1.A | 29.17 | monomer | HHblits | X-ray | 1.95Å | 0.32 | 0.05 | Nicotinamide riboside kinase 1 |
| 2qt0.1.A | 29.17 | monomer | HHblits | X-ray | 1.92Å | 0.32 | 0.05 | Nicotinamide riboside kinase 1 |
| 1gky.1.A | 20.00 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.06 | GUANYLATE KINASE |
| 3h86.2.A | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.05 | Adenylate kinase |
| 3h86.1.B | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.05 | Adenylate kinase |
| 3h86.1.C | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.05 | Adenylate kinase |
| 3h86.1.A | 20.83 | homo-trimer | HHblits | X-ray | 2.50Å | 0.31 | 0.05 | Adenylate kinase |
| 3iim.1.A | 29.17 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.05 | Coilin-interacting nuclear ATPase protein |
| 2anb.1.A | 24.00 | homo-hexamer | HHblits | X-ray | 2.90Å | 0.28 | 0.06 | Guanylate kinase |
| 2anc.1.A | 24.00 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.28 | 0.06 | Guanylate kinase |
| 2anc.1.C | 24.00 | homo-hexamer | HHblits | X-ray | 3.20Å | 0.28 | 0.06 | Guanylate kinase |
| 2an9.1.A | 24.00 | homo-hexamer | HHblits | X-ray | 2.35Å | 0.28 | 0.06 | Guanylate kinase |
| 2an9.1.B | 24.00 | homo-hexamer | HHblits | X-ray | 2.35Å | 0.28 | 0.06 | Guanylate kinase |
| 2f3t.1.D | 24.00 | homo-hexamer | HHblits | X-ray | 3.16Å | 0.28 | 0.06 | Guanylate kinase |
| 4e22.1.A | 29.17 | monomer | HHblits | X-ray | 2.32Å | 0.31 | 0.05 | Cytidylate kinase |
| 1kag.1.A | 25.00 | monomer | HHblits | X-ray | 2.05Å | 0.31 | 0.05 | Shikimate kinase I |
| 1kag.2.A | 25.00 | monomer | HHblits | X-ray | 2.05Å | 0.31 | 0.05 | Shikimate kinase I |
| 3hr7.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.05 | Shikimate kinase |
| 3hr7.1.B | 25.00 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.05 | Shikimate kinase |
| 1zui.1.A | 25.00 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.05 | Shikimate kinase |
| 1zuh.1.A | 25.00 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.05 | Shikimate kinase |
| 3muf.1.A | 25.00 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.05 | Shikimate kinase |
| 1m7h.4.B | 30.43 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.34 | 0.05 | Adenylylsulfate kinase |
| 1d6j.1.B | 30.43 | homo-dimer | HHblits | X-ray | 2.00Å | 0.34 | 0.05 | ADENOSINE-5'PHOSPHOSULFATE KINASE |
| 1d6j.1.A | 30.43 | homo-dimer | HHblits | X-ray | 2.00Å | 0.34 | 0.05 | ADENOSINE-5'PHOSPHOSULFATE KINASE |
| 1m7h.4.C | 30.43 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.34 | 0.05 | Adenylylsulfate kinase |
| 1m7g.5.A | 30.43 | homo-tetramer | HHblits | X-ray | 1.43Å | 0.34 | 0.05 | Adenylylsulfate kinase |
| 2h92.1.A | 30.43 | monomer | HHblits | X-ray | 2.30Å | 0.34 | 0.05 | Cytidylate kinase |
| 3mrs.1.A | 25.00 | monomer | HHblits | X-ray | 2.40Å | 0.31 | 0.05 | Shikimate kinase |
| 3tau.1.A | 16.00 | homo-dimer | HHblits | X-ray | 2.05Å | 0.27 | 0.06 | Guanylate kinase |
| 3tau.1.B | 16.00 | homo-dimer | HHblits | X-ray | 2.05Å | 0.27 | 0.06 | Guanylate kinase |
| 2yvu.1.A | 26.09 | homo-dimer | HHblits | X-ray | 2.10Å | 0.34 | 0.05 | Probable adenylyl-sulfate kinase |
| 2yvu.1.B | 26.09 | homo-dimer | HHblits | X-ray | 2.10Å | 0.34 | 0.05 | Probable adenylyl-sulfate kinase |
| 2uvq.1.A | 20.83 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.30 | 0.05 | URIDINE-CYTIDINE KINASE 1 |
| 3umf.1.A | 25.00 | monomer | HHblits | X-ray | 2.05Å | 0.30 | 0.05 | Adenylate kinase |
| 3trf.1.A | 36.36 | homo-dimer | HHblits | X-ray | 2.60Å | 0.37 | 0.05 | Shikimate kinase |
| 1uky.1.A | 29.17 | monomer | HHblits | X-ray | 2.13Å | 0.30 | 0.05 | URIDYLATE KINASE |
| 4eaq.1.A | 30.43 | homo-dimer | HHblits | X-ray | 1.85Å | 0.33 | 0.05 | Thymidylate kinase |
| 4dwj.1.B | 30.43 | homo-dimer | HHblits | X-ray | 2.74Å | 0.33 | 0.05 | Thymidylate kinase |
| 2ccg.1.A | 30.43 | homo-dimer | HHblits | X-ray | 2.30Å | 0.33 | 0.05 | THYMIDYLATE KINASE |
| 2shk.1.A | 30.43 | homo-dimer | HHblits | X-ray | 2.60Å | 0.33 | 0.05 | SHIKIMATE KINASE |
| 2shk.1.B | 30.43 | homo-dimer | HHblits | X-ray | 2.60Å | 0.33 | 0.05 | SHIKIMATE KINASE |
| 4loa.1.A | 26.09 | homo-dimer | HHblits | X-ray | 1.82Å | 0.33 | 0.05 | De-novo design amidase |
| 4loa.1.B | 26.09 | homo-dimer | HHblits | X-ray | 1.82Å | 0.33 | 0.05 | De-novo design amidase |
| 1ko5.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.28Å | 0.33 | 0.05 | Gluconate kinase |
| 1knq.1.B | 21.74 | homo-dimer | HHblits | X-ray | 2.00Å | 0.33 | 0.05 | Gluconate kinase |
| 1ko8.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.05 | Gluconate kinase |
| 1ko8.1.B | 21.74 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.05 | Gluconate kinase |
| 1ko1.1.B | 21.74 | homo-dimer | HHblits | X-ray | 2.09Å | 0.33 | 0.05 | Gluconate kinase |
| 1knq.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.00Å | 0.33 | 0.05 | Gluconate kinase |
| 3vaa.1.A | 30.43 | monomer | HHblits | X-ray | 1.70Å | 0.33 | 0.05 | Shikimate kinase |
| 3vaa.2.A | 30.43 | monomer | HHblits | X-ray | 1.70Å | 0.33 | 0.05 | Shikimate kinase |
| 3vaa.3.A | 30.43 | monomer | HHblits | X-ray | 1.70Å | 0.33 | 0.05 | Shikimate kinase |
| 2pt5.2.A | 36.36 | monomer | HHblits | X-ray | 2.10Å | 0.36 | 0.05 | Shikimate kinase |
| 2pt5.1.A | 36.36 | monomer | HHblits | X-ray | 2.10Å | 0.36 | 0.05 | Shikimate kinase |
| 2pt5.3.A | 36.36 | monomer | HHblits | X-ray | 2.10Å | 0.36 | 0.05 | Shikimate kinase |
| 2pt5.4.A | 36.36 | monomer | HHblits | X-ray | 2.10Å | 0.36 | 0.05 | Shikimate kinase |
| 2gsd.1.A | 25.00 | homo-dimer | HHblits | X-ray | 1.95Å | 0.29 | 0.05 | NAD-dependent formate dehydrogenase |
| 3ake.1.A | 21.74 | monomer | HHblits | X-ray | 1.50Å | 0.32 | 0.05 | Cytidylate kinase |
| 3akc.1.A | 21.74 | monomer | HHblits | X-ray | 1.65Å | 0.32 | 0.05 | Cytidylate kinase |
| 3w8n.1.A | 21.74 | monomer | HHblits | X-ray | 2.20Å | 0.32 | 0.05 | Cytidylate kinase |
| 3w90.1.A | 21.74 | monomer | HHblits | X-ray | 1.65Å | 0.32 | 0.05 | Cytidylate kinase |
| 5jzv.1.A | 30.43 | monomer | HHblits | X-ray | 2.07Å | 0.32 | 0.05 | Adenylate kinase isoenzyme 6 |
| 4xcq.1.A | 15.38 | monomer | HHblits | X-ray | 2.39Å | 0.23 | 0.06 | TubZ |
| 2jat.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.60Å | 0.36 | 0.05 | DEOXYGUANOSINE KINASE |
| 2jas.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.70Å | 0.36 | 0.05 | DEOXYGUANOSINE KINASE |
| 1z83.1.A | 20.83 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.05 | Adenylate kinase 1 |
| 2c95.1.A | 20.83 | monomer | HHblits | X-ray | 1.71Å | 0.29 | 0.05 | ADENYLATE KINASE 1 |
| 1via.1.A | 26.09 | homo-dimer | HHblits | X-ray | 1.57Å | 0.32 | 0.05 | shikimate kinase |
| 4i1v.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.60Å | 0.35 | 0.05 | Dephospho-CoA kinase |
| 4i1u.1.B | 31.82 | homo-dimer | HHblits | X-ray | 2.05Å | 0.35 | 0.05 | Dephospho-CoA kinase |
| 4i1u.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.05Å | 0.35 | 0.05 | Dephospho-CoA kinase |
| 2ar7.1.A | 12.50 | monomer | HHblits | X-ray | 2.15Å | 0.29 | 0.05 | Adenylate kinase 4 |
| 2ar7.2.A | 12.50 | monomer | HHblits | X-ray | 2.15Å | 0.29 | 0.05 | Adenylate kinase 4 |
| 2bbw.1.A | 12.50 | monomer | HHblits | X-ray | 2.05Å | 0.29 | 0.05 | adenylate kinase 4, AK4 |
| 2bbw.2.A | 12.50 | monomer | HHblits | X-ray | 2.05Å | 0.29 | 0.05 | adenylate kinase 4, AK4 |
| 3adk.1.A | 20.83 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.05 | ADENYLATE KINASE |
| 2bwj.1.A | 20.83 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.05 | ADENYLATE KINASE 5 |
| 2bwj.1.B | 20.83 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.05 | ADENYLATE KINASE 5 |
| 2bwj.2.B | 20.83 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.05 | ADENYLATE KINASE 5 |
| 3n2e.1.A | 26.09 | monomer | HHblits | X-ray | 2.53Å | 0.31 | 0.05 | Shikimate kinase |
| 1e6c.1.A | 26.09 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2ak3.1.A | 17.39 | monomer | HHblits | X-ray | 1.85Å | 0.31 | 0.05 | ADENYLATE KINASE ISOENZYME-3 |
| 2ak3.2.A | 17.39 | monomer | HHblits | X-ray | 1.85Å | 0.31 | 0.05 | ADENYLATE KINASE ISOENZYME-3 |
| 1we2.1.A | 21.74 | monomer | HHblits | X-ray | 2.30Å | 0.31 | 0.05 | Shikimate kinase |
| 1zyu.1.A | 21.74 | monomer | HHblits | X-ray | 2.90Å | 0.31 | 0.05 | Shikimate kinase |
| 3baf.1.A | 21.74 | monomer | HHblits | X-ray | 2.25Å | 0.31 | 0.05 | Shikimate kinase |
| 2g1j.1.A | 21.74 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.05 | Shikimate kinase |
| 2g1j.2.A | 21.74 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.05 | Shikimate kinase |
| 2dfn.1.A | 21.74 | monomer | HHblits | X-ray | 1.93Å | 0.31 | 0.05 | Shikimate kinase |
| 2dft.1.A | 21.74 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.05 | Shikimate kinase |
| 2dft.1.B | 21.74 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.05 | Shikimate kinase |
| 2dft.1.C | 21.74 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.31 | 0.05 | Shikimate kinase |
| 4bqs.2.A | 21.74 | monomer | HHblits | X-ray | 2.15Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 4bqs.3.A | 21.74 | monomer | HHblits | X-ray | 2.15Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2iys.1.A | 21.74 | monomer | HHblits | X-ray | 1.40Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2iyv.1.A | 21.74 | monomer | HHblits | X-ray | 1.35Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2iyx.1.A | 21.74 | monomer | HHblits | X-ray | 1.49Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2iyq.1.A | 21.74 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2iyr.1.A | 21.74 | monomer | HHblits | X-ray | 1.98Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 2iyr.2.A | 21.74 | monomer | HHblits | X-ray | 1.98Å | 0.31 | 0.05 | SHIKIMATE KINASE |
| 4eun.1.A | 21.74 | homo-dimer | HHblits | X-ray | 1.60Å | 0.30 | 0.05 | thermoresistant glucokinase |
| 3fn4.1.A | 26.09 | homo-dimer | HHblits | X-ray | 1.96Å | 0.30 | 0.05 | NAD-dependent formate dehydrogenase |
| 3cm0.1.A | 21.74 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.05 | Adenylate kinase |
| 2cdn.1.A | 21.74 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.05 | ADENYLATE KINASE |
| 4f4j.1.A | 21.74 | homo-dimer | HHblits | X-ray | 2.45Å | 0.30 | 0.05 | Guanylate kinase |
| 4f4j.1.B | 21.74 | homo-dimer | HHblits | X-ray | 2.45Å | 0.30 | 0.05 | Guanylate kinase |
| 2bdt.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.40Å | 0.33 | 0.05 | BH3686 |
| 2jcs.1.A | 17.39 | homo-dimer | HHblits | X-ray | 2.50Å | 0.30 | 0.05 | DEOXYNUCLEOSIDE KINASE |
| 2grj.1.A | 33.33 | monomer | HHblits | X-ray | 2.60Å | 0.37 | 0.05 | Dephospho-CoA kinase |
| 1znx.1.A | 13.04 | monomer | HHblits | X-ray | 2.35Å | 0.30 | 0.05 | Guanylate kinase |
| 4ttq.1.A | 28.57 | monomer | HHblits | X-ray | 2.20Å | 0.37 | 0.05 | Dephospho-CoA kinase |
| 4ttp.1.A | 28.57 | monomer | HHblits | X-ray | 2.20Å | 0.37 | 0.05 | Dephospho-CoA kinase |
| 2qg6.1.A | 31.82 | monomer | HHblits | X-ray | 1.50Å | 0.33 | 0.05 | Nicotinamide riboside kinase 1 |
| 1uf9.1.A | 33.33 | monomer | HHblits | X-ray | 2.80Å | 0.36 | 0.05 | TT1252 protein |
| 1uf9.2.A | 33.33 | monomer | HHblits | X-ray | 2.80Å | 0.36 | 0.05 | TT1252 protein |
| 1uf9.3.A | 33.33 | monomer | HHblits | X-ray | 2.80Å | 0.36 | 0.05 | TT1252 protein |
| 2if2.1.A | 28.57 | monomer | HHblits | X-ray | 3.00Å | 0.36 | 0.05 | Dephospho-CoA kinase |
| 2if2.2.A | 28.57 | monomer | HHblits | X-ray | 3.00Å | 0.36 | 0.05 | Dephospho-CoA kinase |
| 2if2.3.A | 28.57 | monomer | HHblits | X-ray | 3.00Å | 0.36 | 0.05 | Dephospho-CoA kinase |
| 2ukd.1.A | 17.39 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.05 | URIDYLMONOPHOSPHATE/CYTIDYLMONOPHOSPHATE KINASE |
| 1uke.1.A | 17.39 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.05 | URIDYLMONOPHOSPHATE/CYTIDYLMONOPHOSPHATE KINASE |
| 1s4q.1.A | 18.18 | monomer | HHblits | X-ray | 2.16Å | 0.32 | 0.05 | Guanylate kinase |
| 2axp.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.50Å | 0.32 | 0.05 | hypothetical protein BSU20280 |
| 3kb2.1.B | 31.82 | homo-dimer | HHblits | X-ray | 2.20Å | 0.32 | 0.05 | SPBc2 prophage-derived uncharacterized protein yorR |
| 3kb2.1.A | 31.82 | homo-dimer | HHblits | X-ray | 2.20Å | 0.32 | 0.05 | SPBc2 prophage-derived uncharacterized protein yorR |
| 1zak.1.A | 18.18 | homo-dimer | HHblits | X-ray | 3.50Å | 0.32 | 0.05 | ADENYLATE KINASE |
| 3adb.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.05 | L-seryl-tRNA(Sec) kinase |
| 3adc.1.B | 27.27 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.05 | L-seryl-tRNA(Sec) kinase |
| 3adc.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.05 | L-seryl-tRNA(Sec) kinase |
| 3adb.1.B | 27.27 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.05 | L-seryl-tRNA(Sec) kinase |
| 3add.1.B | 27.27 | homo-dimer | HHblits | X-ray | 2.40Å | 0.31 | 0.05 | L-seryl-tRNA(Sec) kinase |
| 2vp9.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.90Å | 0.31 | 0.05 | DEOXYNUCLEOSIDE KINASE |
| 2vp4.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.05 | DEOXYNUCLEOSIDE KINASE |
| 2vp5.1.B | 18.18 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.05 | DEOXYNUCLEOSIDE KINASE |
| 3a1w.1.A | 21.74 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.05 | Iron(II) transport protein B |
| 4np6.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.05 | Adenylate kinase |
| 4np6.1.B | 18.18 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.05 | Adenylate kinase |
| 3fdi.1.A | 17.39 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.27 | 0.05 | uncharacterized protein |
| 1z8f.1.A | 13.64 | monomer | HHblits | X-ray | 2.50Å | 0.30 | 0.05 | Guanylate kinase |
| 2d2e.1.A | 22.73 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.05 | SufC protein |
| 1z6g.1.A | 18.18 | monomer | HHblits | X-ray | 2.18Å | 0.29 | 0.05 | guanylate kinase |
| 3ney.2.A | 13.64 | monomer | HHblits | X-ray | 2.26Å | 0.29 | 0.05 | 55 kDa erythrocyte membrane protein |
| 3ney.1.A | 13.64 | monomer | HHblits | X-ray | 2.26Å | 0.29 | 0.05 | 55 kDa erythrocyte membrane protein |
| 1tev.1.A | 18.18 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.05 | UMP-CMP kinase |
| 1zmx.1.A | 19.05 | homo-dimer | HHblits | X-ray | 3.10Å | 0.32 | 0.05 | Deoxynucleoside kinase |
| 1zm7.1.A | 19.05 | homo-dimer | HHblits | X-ray | 2.20Å | 0.32 | 0.05 | Deoxynucleoside kinase |
| 2j41.1.B | 18.18 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.05 | GUANYLATE KINASE |
| 2j41.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.05 | GUANYLATE KINASE |
| 1p4s.1.A | 23.81 | monomer | HHblits | NMR | NA | 0.31 | 0.05 | Adenylate kinase |
| 3hdt.1.A | 18.18 | homo-dimer | HHblits | X-ray | 2.79Å | 0.27 | 0.05 | putative kinase |
| 3hdt.1.B | 18.18 | homo-dimer | HHblits | X-ray | 2.79Å | 0.27 | 0.05 | putative kinase |
| 4qbh.1.A | 14.29 | monomer | HHblits | X-ray | 1.67Å | 0.30 | 0.05 | Adenylate kinase |
| 4qbh.2.A | 14.29 | monomer | HHblits | X-ray | 1.67Å | 0.30 | 0.05 | Adenylate kinase |
| 1kgd.1.A | 20.00 | monomer | HHblits | X-ray | 1.31Å | 0.33 | 0.05 | PERIPHERAL PLASMA MEMBRANE CASK |
| 2aky.1.A | 19.05 | monomer | HHblits | X-ray | 1.96Å | 0.29 | 0.05 | ADENYLATE KINASE |
| 4zo4.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.57Å | 0.33 | 0.05 | Dephospho-CoA kinase |
| 4zo4.1.B | 25.00 | homo-tetramer | HHblits | X-ray | 2.57Å | 0.33 | 0.05 | Dephospho-CoA kinase |
| 2ak2.1.A | 19.05 | monomer | HHblits | X-ray | 2.10Å | 0.29 | 0.05 | ADENYLATE KINASE ISOENZYME-2 |
| 3a1v.2.A | 23.81 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.05 | Iron(II) transport protein B |
| 3a1v.1.A | 23.81 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.05 | Iron(II) transport protein B |
| 3a1s.2.A | 23.81 | monomer | HHblits | X-ray | 1.50Å | 0.28 | 0.05 | Iron(II) transport protein B |
| 3a1u.2.A | 23.81 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.05 | Iron(II) transport protein B |
| 3a1t.1.A | 23.81 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.05 | Iron(II) transport protein B |
| 3hpr.1.A | 20.00 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.05 | Adenylate kinase |
| 2xbl.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 1.62Å | 0.29 | 0.05 | PHOSPHOHEPTOSE ISOMERASE |
| 4jlo.1.A | 20.00 | monomer | HHblits | X-ray | 1.73Å | 0.29 | 0.05 | Adenylate kinase |
| 4jlo.2.A | 20.00 | monomer | HHblits | X-ray | 1.73Å | 0.29 | 0.05 | Adenylate kinase |
| 4ike.1.A | 20.00 | monomer | HHblits | X-ray | 1.48Å | 0.29 | 0.05 | Adenylate kinase |
| 2rh5.2.A | 20.00 | monomer | HHblits | X-ray | 2.48Å | 0.29 | 0.05 | Adenylate kinase |
| 2rh5.3.A | 20.00 | monomer | HHblits | X-ray | 2.48Å | 0.29 | 0.05 | Adenylate kinase |
| 2rgx.1.A | 20.00 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.05 | Adenylate kinase |
| 2rh5.1.A | 20.00 | monomer | HHblits | X-ray | 2.48Å | 0.29 | 0.05 | Adenylate kinase |
| 4ike.2.A | 20.00 | monomer | HHblits | X-ray | 1.48Å | 0.29 | 0.05 | Adenylate kinase |
| 3sr0.2.A | 20.00 | monomer | HHblits | X-ray | 1.56Å | 0.29 | 0.05 | Adenylate kinase |
| 4cf7.1.A | 20.00 | monomer | HHblits | X-ray | 1.59Å | 0.29 | 0.05 | ADENYLATE KINASE |
| 4cf7.2.A | 20.00 | monomer | HHblits | X-ray | 1.59Å | 0.29 | 0.05 | ADENYLATE KINASE |
| 1zp6.1.A | 21.05 | homo-dimer | HHblits | X-ray | 3.20Å | 0.33 | 0.04 | hypothetical protein Atu3015 |
| 5g3z.1.A | 15.00 | monomer | HHblits | X-ray | 1.89Å | 0.29 | 0.05 | ADENYLATE KINSE |
| 4mkf.1.A | 15.00 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.05 | Adenylate kinase |
| 4mkf.2.A | 15.00 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.05 | Adenylate kinase |
| 1p3j.1.A | 15.00 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.05 | Adenylate kinase |
| 1b0z.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.04 | PROTEIN (PHOSPHOGLUCOSE ISOMERASE) |
| 2q8n.1.A | 31.25 | homo-hexamer | HHblits | X-ray | 1.82Å | 0.34 | 0.04 | Glucose-6-phosphate isomerase |
| 1rcu.1.A | 25.00 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.04 | conserved hypothetical protein VT76 |