 |  | SWISS-MODEL Homology Modelling Report |
Model Building Report
This document lists the results for the homology modelling project "C9BGP5" submitted to SWISS-MODEL workspace on Oct. 26, 2017, 8:43 p.m..The submitted primary amino acid sequence is given in Table T1.
If you use any results in your research, please cite the relevant publications:
Marco Biasini; Stefan Bienert; Andrew Waterhouse; Konstantin Arnold; Gabriel Studer; Tobias Schmidt; Florian Kiefer; Tiziano Gallo Cassarino; Martino Bertoni; Lorenza Bordoli; Torsten Schwede. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research (1 July 2014) 42 (W1): W252-W258; doi: 10.1093/nar/gku340.Arnold, K., Bordoli, L., Kopp, J. and Schwede, T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195-201.
Benkert, P., Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343-350
Results
The SWISS-MODEL template library (SMTL version 2017-10-23, PDB release 2017-10-13) was searched with Blast (Altschul et al., 1997) and HHBlits (Remmert, et al., 2011) for evolutionary related structures matching the target sequence in Table T1. For details on the template search, see Materials and Methods. Overall 1093 templates were found (Table T2).
Models
The following model was built (see Materials and Methods "Model Building"):
Model #01 | File | Built with | Oligo-State | Ligands | GMQE | QMEAN |
|---|---|---|---|---|---|---|
 | PDB | ProMod3 Version 1.1.0. | MONOMER (matching prediction) | None | 0.69 | -2.02 |
|  |  |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 1f0k.1.A | 33.43 | homo-dimer | BLAST | X-ray | 1.90Å | 0.38 | 2 - 361 | 0.93 | UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE |
| Ligand | Added to Model | Description |
|---|---|---|
| SO4 | ✕ - Not biologically relevant. | SULFATE ION |
Target MKILVTGGGTGGHIYPALAFVNYVKTQVPNAEFMYVGAKRGLENKIVPDTGMPFHTLEIQGFKRK-ISMHNIKTIRLFLK
1f0k.1.A -RLMVMAGGTGGHVFPGLAVAHHLMAQ--GWQVRWLGTADRMEADLVPKHGIEIDFIRISGLRGKGIKALIAAPLRIF-N
Target SIRQAKKILSDFQPDIVIGTGGYVSGAVVYAASKMGIPTIIHEQNSVPGVTNKFLSRYVDRIALSFKDAAPFFPENKSVL
1f0k.1.A AWRQARAIMKAYKPDVVLGMGGYVSGPGGLAAWSLGIPVVLHEQNGIAGLTNKWLAKIATKVMQAFPGA---FPNAE--V
Target VGNPRAQEVADTKKSDILKTFGLDPEKK--------TVLIFGGSQGALKINQAVSVFLSEYIPSSYQVLYASGERYYEEI
1f0k.1.A VGNP--------VRTDVLAL--PLPQQRLAGREGPVRVLVVGGSQGARILNQTMPQVAAK-LGDSVTIWHQSGKGSQQSV
Target LNKINPEIQ-NISVQPYIKNMAEVMASSDLLVGRAGATSIAEFTALGLPAVLVPSPYVTNDHQTKNAMSLVHAGAAKMIA
1f0k.1.A EQAYAEAGQPQHKVTEFIDDMAAAYAWADVVVCRSGALTVSEIAAAGLPALFVPFQH-KDRQQYWNALPLEKAGAAKIIE
Target DNELTGESLSQTINEIMDDEELQKQMCRASKEQGIPDASKRLYDVVNQIIKK
1f0k.1.A QPQLSVDAVANTLAGWSRETLL--TMAERARAASIPDATERVANEVSRVAR-
Materials and Methods
Template Search
Template search with Blast and HHBlits has been performed against the SWISS-MODEL template library (SMTL, last update: 2017-10-23, last included PDB release: 2017-10-13).
The target sequence was searched with BLAST (Altschul et al., 1997) against the primary amino acid sequence contained in the SMTL. A total of 10 templates were found.
An initial HHblits profile has been built using the procedure outlined in (Remmert, et al., 2011), followed by 1 iteration of HHblits against NR20. The obtained profile has then be searched against all profiles of the SMTL. A total of 1086 templates were found.
Template Selection
For each identified template, the template's quality has been predicted from features of the target-template alignment. The templates with the highest quality have then been selected for model building.
Model Building
Models are built based on the target-template alignment using ProMod3. Coordinates which are conserved between the target and the template are copied from the template to the model. Insertions and deletions are remodelled using a fragment library. Side chains are then rebuilt. Finally, the geometry of the resulting model is regularized by using a force field. In case loop modelling with ProMod3 fails, an alternative model is built with PROMOD-II (Guex, et al., 1997).
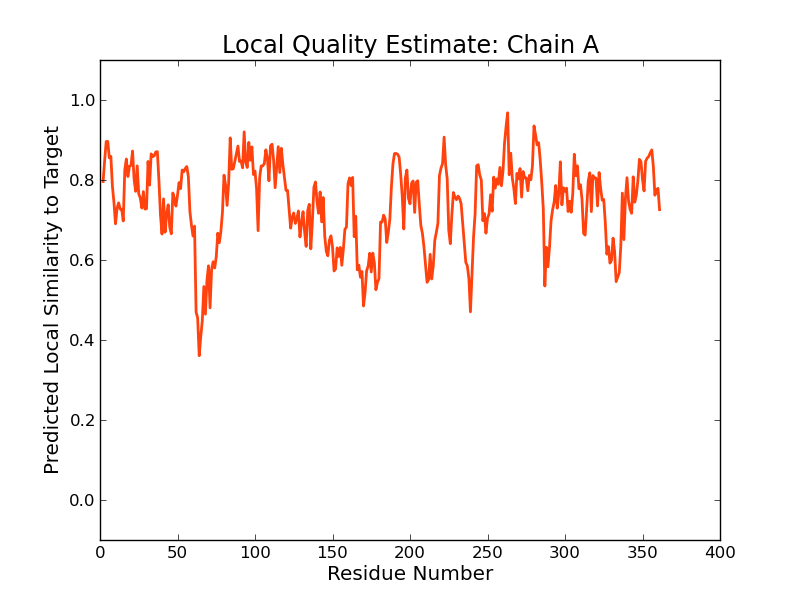
Model Quality Estimation
The global and per-residue model quality has been assessed using the QMEAN scoring function (Benkert, et al., 2011) . For improved performance, weights of the individual QMEAN terms have been trained specifically for SWISS-MODEL.
Ligand Modelling
Ligands present in the template structure are transferred by homology to the model when the following criteria are met (Gallo -Casserino, to be published): (a) The ligands are annotated as biologically relevant in the template library, (b) the ligand is in contact with the model, (c) the ligand is not clashing with the protein, (d) the residues in contact with the ligand are conserved between the target and the template. If any of these four criteria is not satisfied, a certain ligand will not be included in the model. The model summary includes information on why and which ligand has not been included.
Oligomeric State Conservation
Homo-oligomeric structure of the target protein is predicted based on the analysis of pairwise interfaces of the identified template structures. For each relevant interface between polypeptide chains (interfaces with more than 10 residue-residue interactions), the QscoreOligomer (Mariani et al., 2011) is predicted from features such as similarity to target and frequency of observing this interface in the identified templates (Kiefer, Bertoni, Biasini, to be published). The prediction is performed with a random forest regressor using these features as input parameters to predict the probability of conservation for each interface. The QscoreOligomer of the whole complex is then calculated as the weight-averaged QscoreOligomer of the interfaces. The oligomeric state of the target is predicted to be the same as in the template when QscoreOligomer is predicted to be higher or equal to 0.5.
References
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res, 25, 3389-3402.
Remmert, M., Biegert, A., Hauser, A. and Soding, J. (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods, 9, 173-175.
Guex, N. and Peitsch, M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714-2723.
Sali, A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol, 234, 779-815.
Benkert, P., Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343-350.
Mariani, V., Kiefer, F., Schmidt, T., Haas, J. and Schwede, T. (2011) Assessment of template based protein structure predictions in CASP9. Proteins, 79 Suppl 10, 37-58.
Table T1:
Primary amino acid sequence for which templates were searched and models were built.
GYVSGAVVYAASKMGIPTIIHEQNSVPGVTNKFLSRYVDRIALSFKDAAPFFPENKSVLVGNPRAQEVADTKKSDILKTFGLDPEKKTVLIFGGSQGALK
INQAVSVFLSEYIPSSYQVLYASGERYYEEILNKINPEIQNISVQPYIKNMAEVMASSDLLVGRAGATSIAEFTALGLPAVLVPSPYVTNDHQTKNAMSL
VHAGAAKMIADNELTGESLSQTINEIMDDEELQKQMCRASKEQGIPDASKRLYDVVNQIIKK
Table T2:
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Coverage | Description |
|---|---|---|---|---|---|---|---|---|
| 3s2u.1.A | 32.38 | monomer | BLAST | X-ray | 2.23Å | 0.37 | 0.96 | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| 3s2u.1.A | 31.71 | monomer | HHblits | X-ray | 2.23Å | 0.37 | 0.97 | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| 1f0k.1.A | 30.84 | homo-dimer | HHblits | X-ray | 1.90Å | 0.36 | 0.96 | UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE |
| 1f0k.1.B | 30.84 | homo-dimer | HHblits | X-ray | 1.90Å | 0.36 | 0.96 | UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE |
| 1nlm.1.A | 30.84 | homo-dimer | HHblits | X-ray | 2.50Å | 0.36 | 0.96 | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| 1nlm.1.B | 30.84 | homo-dimer | HHblits | X-ray | 2.50Å | 0.36 | 0.96 | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| 1f0k.1.A | 33.43 | homo-dimer | BLAST | X-ray | 1.90Å | 0.38 | 0.93 | UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE |
| 1f0k.1.B | 33.43 | homo-dimer | BLAST | X-ray | 1.90Å | 0.38 | 0.93 | UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE |
| 1nlm.1.A | 33.43 | homo-dimer | BLAST | X-ray | 2.50Å | 0.38 | 0.93 | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| 1nlm.1.B | 33.43 | homo-dimer | BLAST | X-ray | 2.50Å | 0.38 | 0.93 | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| 4wyi.1.A | 17.87 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.96 | Monogalactosyldiacylglycerol synthase 1, chloroplastic |
| 4x1t.1.A | 17.87 | monomer | HHblits | X-ray | 2.25Å | 0.29 | 0.96 | Monogalactosyldiacylglycerol synthase 1, chloroplastic |
| 3otg.1.A | 16.03 | monomer | HHblits | X-ray | 2.08Å | 0.29 | 0.95 | CalG1 |
| 3oth.1.A | 16.03 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.95 | CalG1 |
| 4fzr.1.A | 18.24 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.94 | SsfS6 |
| 4rie.1.A | 14.41 | homo-dimer | HHblits | X-ray | 2.16Å | 0.29 | 0.94 | Glycosyl transferase homolog |
| 4rie.1.B | 14.41 | homo-dimer | HHblits | X-ray | 2.16Å | 0.29 | 0.94 | Glycosyl transferase homolog |
| 4rif.1.A | 14.41 | homo-dimer | HHblits | X-ray | 1.85Å | 0.29 | 0.94 | Glycosyl transferase homolog |
| 4rif.1.B | 14.41 | homo-dimer | HHblits | X-ray | 1.85Å | 0.29 | 0.94 | Glycosyl transferase homolog |
| 3wc4.1.A | 14.78 | monomer | HHblits | X-ray | 1.85Å | 0.28 | 0.95 | UDP-glucose:anthocyanidin 3-O-glucosyltransferase |
| 4g2t.1.A | 17.94 | homo-dimer | HHblits | X-ray | 2.41Å | 0.29 | 0.94 | SsfS6 |
| 4rii.1.A | 14.12 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.94 | Glycosyl transferase homolog,Glycosyl transferase |
| 4rih.1.A | 14.12 | homo-dimer | HHblits | X-ray | 2.22Å | 0.29 | 0.94 | Glycosyl transferase homolog,Glycosyl transferase |
| 4rig.1.B | 14.12 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.94 | Glycosyl transferase |
| 4rig.1.A | 14.12 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.94 | Glycosyl transferase |
| 3beo.1.A | 17.80 | homo-dimer | HHblits | X-ray | 1.70Å | 0.30 | 0.93 | UDP-N-acetylglucosamine 2-epimerase |
| 4m60.1.A | 18.99 | monomer | HHblits | X-ray | 1.77Å | 0.30 | 0.93 | Oleandomycin glycosyltransferase |
| 4m83.1.A | 19.05 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.93 | Oleandomycin glycosyltransferase |
| 4m83.2.A | 19.05 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.93 | Oleandomycin glycosyltransferase |
| 2iyf.1.A | 18.99 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.93 | OLEANDOMYCIN GLYCOSYLTRANSFERASE |
| 2iyf.2.A | 18.99 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.93 | OLEANDOMYCIN GLYCOSYLTRANSFERASE |
| 2iya.1.A | 17.80 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.93 | OLEANDOMYCIN GLYCOSYLTRANSFERASE |
| 5enz.1.A | 18.15 | homo-dimer | HHblits | X-ray | 1.91Å | 0.29 | 0.93 | UDP-GlcNAc 2-epimerase |
| 5enz.1.B | 18.15 | homo-dimer | HHblits | X-ray | 1.91Å | 0.29 | 0.93 | UDP-GlcNAc 2-epimerase |
| 4rbn.1.A | 13.95 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.27 | 0.95 | Sucrose synthase:Glycosyl transferases group 1 |
| 4rbn.1.B | 13.95 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.27 | 0.95 | Sucrose synthase:Glycosyl transferases group 1 |
| 4rbn.1.C | 13.95 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.27 | 0.95 | Sucrose synthase:Glycosyl transferases group 1 |
| 4rbn.1.D | 13.95 | homo-tetramer | HHblits | X-ray | 3.05Å | 0.27 | 0.95 | Sucrose synthase:Glycosyl transferases group 1 |
| 5dld.1.A | 19.22 | monomer | HHblits | X-ray | 1.45Å | 0.30 | 0.92 | UDP-N-Acetylglucosamine 2-epimerase |
| 2yjn.1.A | 13.82 | hetero-oligomer | HHblits | X-ray | 3.09Å | 0.28 | 0.94 | GLYCOSYLTRANSFERASE |
| 3ia7.1.A | 15.93 | homo-dimer | HHblits | X-ray | 1.91Å | 0.28 | 0.94 | CalG4 |
| 3ia7.1.B | 15.93 | homo-dimer | HHblits | X-ray | 1.91Å | 0.28 | 0.94 | CalG4 |
| 5gl5.1.A | 13.74 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.94 | Sterol 3-beta-glucosyltransferase |
| 5gl5.1.B | 13.74 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.94 | Sterol 3-beta-glucosyltransferase |
| 1f6d.1.B | 16.47 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.92 | UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE |
| 1f6d.1.A | 16.47 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.92 | UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE |
| 5xvm.1.A | 13.74 | monomer | HHblits | X-ray | 2.77Å | 0.27 | 0.94 | Sterol 3-beta-glucosyltransferase |
| 4amb.1.A | 14.84 | homo-dimer | HHblits | X-ray | 2.62Å | 0.28 | 0.93 | SNOGD |
| 4amg.1.A | 14.84 | homo-dimer | HHblits | X-ray | 2.59Å | 0.28 | 0.93 | SNOGD |
| 4amg.1.B | 14.84 | homo-dimer | HHblits | X-ray | 2.59Å | 0.28 | 0.93 | SNOGD |
| 4an4.1.A | 14.84 | homo-dimer | HHblits | X-ray | 2.70Å | 0.28 | 0.93 | GLYCOSYL TRANSFERASE |
| 4an4.1.B | 14.84 | homo-dimer | HHblits | X-ray | 2.70Å | 0.28 | 0.93 | GLYCOSYL TRANSFERASE |
| 4an4.2.A | 14.84 | homo-dimer | HHblits | X-ray | 2.70Å | 0.28 | 0.93 | GLYCOSYL TRANSFERASE |
| 1vgv.1.B | 16.17 | homo-dimer | HHblits | X-ray | 2.31Å | 0.29 | 0.92 | UDP-N-acetylglucosamine 2-epimerase |
| 1vgv.1.A | 16.17 | homo-dimer | HHblits | X-ray | 2.31Å | 0.29 | 0.92 | UDP-N-acetylglucosamine 2-epimerase |
| 1vgv.2.B | 16.17 | homo-dimer | HHblits | X-ray | 2.31Å | 0.29 | 0.92 | UDP-N-acetylglucosamine 2-epimerase |
| 3d1j.1.A | 13.95 | monomer | HHblits | X-ray | 3.00Å | 0.27 | 0.95 | Glycogen synthase |
| 3ot5.1.A | 14.50 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.93 | UDP-N-acetylglucosamine 2-epimerase |
| 3ot5.2.A | 14.50 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.93 | UDP-N-acetylglucosamine 2-epimerase |
| 3wad.1.A | 13.82 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.94 | Glycosyltransferase |
| 1v4v.1.A | 17.31 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.93 | UDP-N-Acetylglucosamine 2-Epimerase |
| 1v4v.1.B | 17.31 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.93 | UDP-N-Acetylglucosamine 2-Epimerase |
| 3l01.1.A | 12.24 | monomer | HHblits | X-ray | 2.60Å | 0.26 | 0.95 | GlgA glycogen synthase |
| 3dzc.1.A | 16.77 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.92 | UDP-N-acetylglucosamine 2-epimerase |
| 3dzc.1.B | 16.77 | homo-dimer | HHblits | X-ray | 2.35Å | 0.28 | 0.92 | UDP-N-acetylglucosamine 2-epimerase |
| 3guh.1.A | 13.70 | monomer | HHblits | X-ray | 2.79Å | 0.26 | 0.95 | Glycogen synthase |
| 3mbo.1.A | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.1.B | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.2.A | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.2.B | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.3.A | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.3.B | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.4.A | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 3mbo.4.B | 13.91 | homo-dimer | HHblits | X-ray | 3.31Å | 0.27 | 0.93 | Glycosyltransferase, group 1 family |
| 5du2.1.A | 15.73 | monomer | HHblits | X-ray | 2.70Å | 0.28 | 0.93 | EspG2 glycosyltransferase |
| 5du2.2.A | 15.73 | monomer | HHblits | X-ray | 2.70Å | 0.28 | 0.93 | EspG2 glycosyltransferase |
| 1rzu.1.A | 12.83 | monomer | HHblits | X-ray | 2.30Å | 0.26 | 0.95 | Glycogen synthase 1 |
| 1rzu.2.A | 12.83 | monomer | HHblits | X-ray | 2.30Å | 0.26 | 0.95 | Glycogen synthase 1 |
| 3cop.1.A | 13.41 | monomer | HHblits | X-ray | 2.30Å | 0.26 | 0.95 | Glycogen synthase |
| 3fro.1.A | 12.87 | homo-trimer | HHblits | X-ray | 2.50Å | 0.26 | 0.94 | GlgA glycogen synthase |
| 3fro.1.B | 12.87 | homo-trimer | HHblits | X-ray | 2.50Å | 0.26 | 0.94 | GlgA glycogen synthase |
| 3fro.1.C | 12.87 | homo-trimer | HHblits | X-ray | 2.50Å | 0.26 | 0.94 | GlgA glycogen synthase |
| 4hwg.1.A | 17.22 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.29 | 0.91 | UDP-N-acetylglucosamine 2-epimerase |
| 2p6p.1.A | 15.32 | homo-dimer | HHblits | X-ray | 1.88Å | 0.28 | 0.92 | Glycosyl transferase |
| 2p6p.1.B | 15.32 | homo-dimer | HHblits | X-ray | 1.88Å | 0.28 | 0.92 | Glycosyl transferase |
| 5d00.1.A | 14.79 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.93 | N-acetyl-alpha-D-glucosaminyl L-malate synthase |
| 1o6c.1.A | 15.22 | homo-dimer | HHblits | X-ray | 2.90Å | 0.28 | 0.93 | UDP-N-acetylglucosamine 2-epimerase |
| 1o6c.1.B | 15.22 | homo-dimer | HHblits | X-ray | 2.90Å | 0.28 | 0.93 | UDP-N-acetylglucosamine 2-epimerase |
| 4fkz.1.A | 15.22 | homo-dimer | HHblits | X-ray | 1.69Å | 0.28 | 0.93 | UDP-N-acetylglucosamine 2-epimerase |
| 3rsc.1.A | 13.86 | monomer | HHblits | X-ray | 2.19Å | 0.27 | 0.94 | CalG2 |
| 3iaa.1.A | 13.86 | homo-dimer | HHblits | X-ray | 2.51Å | 0.27 | 0.94 | CalG2 |
| 4xsr.1.A | 12.06 | homo-dimer | HHblits | X-ray | 2.39Å | 0.27 | 0.94 | Alr3699 protein |
| 4xsp.1.B | 12.06 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.94 | Alr3699 protein |
| 4xso.1.A | 12.06 | homo-dimer | HHblits | X-ray | 2.01Å | 0.27 | 0.94 | Alr3699 protein |
| 2bis.1.A | 12.61 | monomer | HHblits | X-ray | 2.80Å | 0.26 | 0.94 | GLGA GLYCOGEN SYNTHASE |
| 2bis.2.A | 12.61 | monomer | HHblits | X-ray | 2.80Å | 0.26 | 0.94 | GLGA GLYCOGEN SYNTHASE |
| 2r68.1.A | 12.57 | monomer | HHblits | X-ray | 2.40Å | 0.26 | 0.94 | Glycosyl transferase, group 1 |
| 4lei.1.A | 12.54 | homo-dimer | HHblits | X-ray | 3.15Å | 0.28 | 0.93 | NDP-forosamyltransferase |
| 4lei.1.B | 12.54 | homo-dimer | HHblits | X-ray | 3.15Å | 0.28 | 0.93 | NDP-forosamyltransferase |
| 3c4q.1.A | 9.94 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.94 | Predicted glycosyltransferases |
| 3c48.1.A | 9.94 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.94 | Predicted glycosyltransferases |
| 4zht.1.A | 14.50 | homo-tetramer | HHblits | X-ray | 2.69Å | 0.28 | 0.91 | Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase |
| 1pnv.1.A | 12.65 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.94 | GLYCOSYLTRANSFERASE GTFA |
| 1pnv.1.B | 12.65 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.94 | GLYCOSYLTRANSFERASE GTFA |
| 3h4i.1.A | 15.18 | monomer | HHblits | X-ray | 1.30Å | 0.27 | 0.93 | Glycosyltransferase GtfA, Glycosyltransferase |
| 1iir.1.A | 14.24 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.93 | glycosyltransferase GtfB |
| 2acw.1.A | 17.99 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.91 | triterpene UDP-glucosyl transferase UGT71G1 |
| 2acw.1.B | 17.99 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.91 | triterpene UDP-glucosyl transferase UGT71G1 |
| 2xa9.1.A | 14.11 | monomer | HHblits | X-ray | 2.50Å | 0.27 | 0.92 | TREHALOSE-SYNTHASE TRET |
| 2xa1.1.A | 13.81 | monomer | HHblits | X-ray | 2.47Å | 0.27 | 0.92 | TREHALOSE-SYNTHASE TRET |
| 2x6r.1.A | 13.81 | monomer | HHblits | X-ray | 2.20Å | 0.27 | 0.92 | TREHALOSE-SYNTHASE TRET |
| 2acv.1.A | 17.68 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.91 | triterpene UDP-glucosyl transferase UGT71G1 |
| 2acv.1.B | 17.68 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.91 | triterpene UDP-glucosyl transferase UGT71G1 |
| 3tsa.1.A | 12.57 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.92 | NDP-rhamnosyltransferase |
| 3tsa.1.B | 12.57 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.92 | NDP-rhamnosyltransferase |
| 3oti.1.A | 13.15 | monomer | HHblits | X-ray | 1.60Å | 0.28 | 0.90 | CalG3 |
| 3d0r.1.B | 13.15 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.90 | Protein CalG3 |
| 3d0r.1.A | 13.15 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.90 | Protein CalG3 |
| 3d0q.1.A | 13.15 | homo-dimer | HHblits | X-ray | 2.79Å | 0.28 | 0.90 | Protein CalG3 |
| 2jjm.1.A | 13.33 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.27 | 0.91 | GLYCOSYL TRANSFERASE, GROUP 1 FAMILY PROTEIN |
| 2gek.1.A | 13.90 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.91 | PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) |
| 4nc9.2.A | 12.91 | monomer | HHblits | X-ray | 3.19Å | 0.26 | 0.92 | GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase |
| 4nc9.1.A | 12.91 | monomer | HHblits | X-ray | 3.19Å | 0.26 | 0.92 | GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase |
| 4n9w.1.A | 12.91 | monomer | HHblits | X-ray | 1.94Å | 0.26 | 0.92 | GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase |
| 1rrv.1.A | 12.01 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.92 | GLYCOSYLTRANSFERASE GTFD |
| 3uyl.1.A | 12.35 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.92 | NDP-rhamnosyltransferase |
| 5u6n.2.A | 16.10 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.89 | UDP-glycosyltransferase 74F2 |
| 5u6n.1.A | 16.10 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.89 | UDP-glycosyltransferase 74F2 |
| 5v2k.1.A | 16.10 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.89 | UDP-glycosyltransferase 74F2 |
| 5v2k.2.A | 16.10 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.89 | UDP-glycosyltransferase 74F2 |
| 4neq.1.A | 15.17 | homo-dimer | HHblits | X-ray | 2.85Å | 0.28 | 0.89 | UDP-N-acetylglucosamine 2-epimerase |
| 4nes.1.A | 15.17 | homo-dimer | HHblits | X-ray | 1.42Å | 0.28 | 0.89 | UDP-N-acetylglucosamine 2-epimerase |
| 2c1x.1.A | 14.55 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.89 | UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE |
| 3oka.1.A | 12.08 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.26 | 0.91 | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase |
| 5u6m.1.A | 15.63 | monomer | HHblits | X-ray | 2.57Å | 0.28 | 0.88 | UDP-glycosyltransferase 74F2 |
| 5u6m.2.A | 15.63 | monomer | HHblits | X-ray | 2.57Å | 0.28 | 0.88 | UDP-glycosyltransferase 74F2 |
| 5u6s.1.A | 15.63 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.88 | UDP-glycosyltransferase 74F2 |
| 5u6s.2.A | 15.63 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.88 | UDP-glycosyltransferase 74F2 |
| 3okp.1.A | 12.42 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.91 | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase |
| 2iv7.1.A | 11.21 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.91 | LIPOPOLYSACCHARIDE CORE BIOSYNTHESIS PROTEIN RFAG |
| 5tmd.1.A | 14.69 | monomer | HHblits | X-ray | 2.19Å | 0.28 | 0.88 | Glycosyltransferase, Os79 |
| 5tmb.1.A | 14.69 | monomer | HHblits | X-ray | 2.34Å | 0.28 | 0.88 | Glycosyltransferase, Os79 |
| 2vg8.1.A | 17.67 | monomer | HHblits | X-ray | 1.75Å | 0.29 | 0.88 | HYDROQUINONE GLUCOSYLTRANSFERASE |
| 3oy2.1.A | 11.21 | homo-dimer | HHblits | X-ray | 2.31Å | 0.26 | 0.91 | Glycosyltransferase B736L |
| 2pq6.1.A | 14.73 | monomer | HHblits | X-ray | 2.10Å | 0.28 | 0.88 | UDP-glucuronosyl/UDP-glucosyltransferase |
| 2vsn.1.A | 10.80 | monomer | HHblits | X-ray | 2.75Å | 0.25 | 0.90 | XCOGT |
| 2xgs.1.A | 10.49 | monomer | HHblits | X-ray | 2.39Å | 0.25 | 0.90 | XCOGT |
| 2xgm.2.A | 10.49 | monomer | HHblits | X-ray | 2.55Å | 0.25 | 0.90 | XCOGT |
| 2vsy.1.A | 10.49 | monomer | HHblits | X-ray | 2.10Å | 0.25 | 0.90 | XCC0866 |
| 2vsy.2.A | 10.49 | monomer | HHblits | X-ray | 2.10Å | 0.25 | 0.90 | XCC0866 |
| 3hbf.1.A | 14.29 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.87 | Flavonoid 3-O-glucosyltransferase |
| 3hbj.1.A | 14.29 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.87 | Flavonoid 3-O-glucosyltransferase |
| 4xyw.1.A | 15.48 | monomer | HHblits | X-ray | 2.20Å | 0.27 | 0.86 | O-antigen biosynthesis glycosyltransferase WbnH |
| 2h1f.1.A | 14.29 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.83 | Lipopolysaccharide heptosyltransferase-1 |
| 2h1f.2.A | 14.29 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.83 | Lipopolysaccharide heptosyltransferase-1 |
| 2gt1.1.A | 14.33 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.83 | Lipopolysaccharide heptosyltransferase-1 |
| 2gt1.2.A | 14.33 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.83 | Lipopolysaccharide heptosyltransferase-1 |
| 2xci.1.A | 13.61 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.81 | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE |
| 2iv3.1.A | 11.42 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.80 | GLYCOSYLTRANSFERASE |
| 4w6q.1.A | 13.03 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.78 | glucosyltransferase |
| 4w6q.1.B | 13.03 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.78 | glucosyltransferase |
| 4w6q.1.C | 13.03 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.78 | glucosyltransferase |
| 4w6q.1.D | 13.03 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.78 | glucosyltransferase |
| 1psw.1.A | 16.92 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.72 | ADP-HEPTOSE LPS HEPTOSYLTRANSFERASE II |
| 3q3h.1.A | 10.94 | monomer | HHblits | X-ray | 2.25Å | 0.26 | 0.73 | HMW1C-like glycosyltransferase |
| 3tov.1.A | 14.67 | monomer | HHblits | X-ray | 2.98Å | 0.27 | 0.72 | Glycosyl transferase family 9 |
| 3tov.2.A | 14.67 | monomer | HHblits | X-ray | 2.98Å | 0.27 | 0.72 | Glycosyl transferase family 9 |
| 4x6l.1.A | 12.79 | homo-trimer | HHblits | X-ray | 3.19Å | 0.27 | 0.71 | TarM |
| 4x7p.1.A | 12.79 | homo-trimer | HHblits | X-ray | 3.40Å | 0.27 | 0.71 | TarM |
| 4wac.1.A | 13.18 | homo-trimer | HHblits | X-ray | 2.40Å | 0.27 | 0.71 | Glycosyl transferase, group 1 family protein |
| 4wad.1.A | 13.18 | homo-trimer | HHblits | X-ray | 2.80Å | 0.27 | 0.71 | Glycosyl transferase, group 1 family protein |
| 3hbm.1.A | 15.18 | monomer | HHblits | X-ray | 1.80Å | 0.28 | 0.71 | UDP-sugar hydrolase |
| 3hbn.1.A | 15.18 | monomer | HHblits | X-ray | 1.85Å | 0.28 | 0.71 | UDP-sugar hydrolase |
| 3s27.1.A | 12.79 | homo-tetramer | HHblits | X-ray | 2.91Å | 0.27 | 0.71 | Sucrose synthase 1 |
| 3s27.1.B | 12.79 | homo-tetramer | HHblits | X-ray | 2.91Å | 0.27 | 0.71 | Sucrose synthase 1 |
| 4x7r.1.A | 12.02 | monomer | HHblits | X-ray | 2.15Å | 0.27 | 0.71 | TarM |
| 4x7m.2.A | 12.02 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.71 | Uncharacterized protein |
| 4x7m.1.A | 12.02 | monomer | HHblits | X-ray | 2.40Å | 0.27 | 0.71 | Uncharacterized protein |
| 5djs.1.A | 8.46 | monomer | HHblits | X-ray | 2.80Å | 0.25 | 0.72 | Tetratricopeptide TPR_2 repeat protein |
| 4pqg.1.A | 12.20 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 5e9t.1.A | 11.42 | hetero-oligomer | HHblits | X-ray | 2.92Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 5e9t.1.C | 11.42 | hetero-oligomer | HHblits | X-ray | 2.92Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 5e9u.1.A | 11.42 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 5e9u.1.C | 11.42 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 5e9u.2.A | 11.42 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 5e9u.2.C | 11.42 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.70 | Glycosyltransferase Gtf1 |
| 4f96.1.A | 8.56 | homo-dimer | HHblits | X-ray | 2.15Å | 0.25 | 0.71 | VldE |
| 4f9f.1.A | 8.56 | homo-dimer | HHblits | X-ray | 2.81Å | 0.25 | 0.71 | VldE |
| 4f9f.1.B | 8.56 | homo-dimer | HHblits | X-ray | 2.81Å | 0.25 | 0.71 | VldE |
| 4f9f.2.A | 8.56 | homo-dimer | HHblits | X-ray | 2.81Å | 0.25 | 0.71 | VldE |
| 3t5t.1.A | 8.56 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.71 | Putative glycosyltransferase |
| 3t5t.1.B | 8.56 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.71 | Putative glycosyltransferase |
| 5lqd.1.A | 10.55 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.71 | Alpha,alpha-trehalose-phosphate synthase |
| 5lqd.1.B | 10.55 | homo-dimer | HHblits | X-ray | 1.95Å | 0.25 | 0.71 | Alpha,alpha-trehalose-phosphate synthase |
| 3rhz.1.A | 11.69 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.26 | 0.64 | Nucleotide sugar synthetase-like protein |
| 3qkw.1.A | 11.35 | homo-tetramer | HHblits | X-ray | 2.29Å | 0.26 | 0.63 | Nucleotide sugar synthetase-like protein |
| 3cv3.1.A | 12.89 | monomer | HHblits | X-ray | 2.25Å | 0.26 | 0.62 | Glucuronosyltransferase GumK |
| 2q6v.1.A | 12.44 | monomer | HHblits | X-ray | 2.28Å | 0.26 | 0.62 | Glucuronosyltransferase GumK |
| 3l7k.1.A | 11.85 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.27 | 0.58 | Teichoic acid biosynthesis protein F |
| 3l7k.1.D | 11.85 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.27 | 0.58 | Teichoic acid biosynthesis protein F |
| 3l7m.1.A | 12.32 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.27 | 0.58 | Teichoic acid biosynthesis protein F |
| 3vue.1.A | 10.85 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.59 | Granule-bound starch synthase 1, chloroplastic/amyloplastic |
| 3l7i.1.A | 11.85 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.26 | 0.58 | Teichoic acid biosynthesis protein F |
| 4hln.1.A | 9.95 | monomer | HHblits | X-ray | 2.70Å | 0.26 | 0.58 | Starch synthase I |
| 5hut.1.A | 13.11 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.57 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] |
| 5i45.1.A | 10.22 | monomer | HHblits | X-ray | 1.35Å | 0.26 | 0.51 | Glycosyl transferases group 1 family protein |
| 5hvm.1.A | 13.04 | homo-dimer | HHblits | X-ray | 2.82Å | 0.26 | 0.51 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 3t7d.1.A | 9.19 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.51 | Putative glycosyltransferase |
| 2wtx.1.A | 6.99 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.51 | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE [UDP-FORMING] |
| 2wtx.1.B | 6.99 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.51 | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE [UDP-FORMING] |
| 1uqu.1.A | 6.99 | monomer | HHblits | X-ray | 2.00Å | 0.24 | 0.51 | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE |
| 1uqu.2.A | 6.99 | monomer | HHblits | X-ray | 2.00Å | 0.24 | 0.51 | ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE |
| 1gz5.1.A | 6.99 | homo-tetramer | HHblits | X-ray | 2.43Å | 0.24 | 0.51 | ALPHA-TREHALOSE-PHOSPHATE SYNTHASE |
| 5uof.1.A | 8.60 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.51 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 5hxa.1.A | 8.06 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.51 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 5tvg.1.A | 8.65 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.24 | 0.51 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 5tvg.2.A | 8.65 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.24 | 0.51 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 4bfc.1.A | 11.43 | homo-trimer | HHblits | X-ray | 1.70Å | 0.27 | 0.48 | 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE |
| 5lwv.1.A | 14.86 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.48 | Host cell factor 1,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
| 5lvv.1.A | 14.94 | monomer | HHblits | X-ray | 2.54Å | 0.27 | 0.48 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
| 5hgv.1.A | 14.94 | hetero-oligomer | HHblits | X-ray | 2.05Å | 0.27 | 0.48 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
| 4ay5.1.A | 14.94 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.27 | 0.48 | UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYL TRANSFERASE 110 KDA SUBUNIT |
| 4gyw.1.A | 14.94 | hetero-oligomer | HHblits | X-ray | 1.70Å | 0.27 | 0.48 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
| 4n3c.1.A | 14.94 | hetero-oligomer | HHblits | X-ray | 2.55Å | 0.27 | 0.48 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
| 5c1d.1.A | 14.94 | monomer | HHblits | X-ray | 2.05Å | 0.27 | 0.48 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit |
| 5a01.1.A | 12.00 | monomer | HHblits | X-ray | 2.66Å | 0.26 | 0.48 | O-GLYCOSYLTRANSFERASE |
| 5u09.1.A | 12.18 | monomer | HHblits | X-ray | 2.60Å | 0.27 | 0.43 | Cannabinoid receptor 1,GlgA glycogen synthase |
| 4s0v.1.A | 12.18 | monomer | HHblits | X-ray | 2.50Å | 0.27 | 0.43 | Human Orexin receptor type 2 fusion protein to P. abysii Glycogen Synthase |
| 2bfw.1.A | 12.26 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.43 | GLGA GLYCOGEN SYNTHASE |
| 3qhp.1.A | 11.18 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.42 | Type 1 capsular polysaccharide biosynthesis protein J (CapJ) |
| 3qhp.2.A | 11.18 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.42 | Type 1 capsular polysaccharide biosynthesis protein J (CapJ) |
| 2o6l.1.A | 18.06 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.40 | UDP-glucuronosyltransferase 2B7 |
| 2o6l.1.B | 18.06 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.40 | UDP-glucuronosyltransferase 2B7 |
| 5e9t.1.B | 8.67 | hetero-oligomer | HHblits | X-ray | 2.92Å | 0.26 | 0.41 | Glycosyltransferase-stabilizing protein Gtf2 |
| 5e9t.1.D | 8.67 | hetero-oligomer | HHblits | X-ray | 2.92Å | 0.26 | 0.41 | Glycosyltransferase-stabilizing protein Gtf2 |
| 2jzc.1.A | 20.44 | monomer | HHblits | NMR | NA | 0.30 | 0.38 | UDP-N-acetylglucosamine transferase subunit ALG13 |
| 2ks6.1.A | 19.85 | monomer | HHblits | NMR | NA | 0.30 | 0.38 | UDP-N-acetylglucosamine transferase subunit ALG13 |
| 2x0d.1.A | 11.51 | homo-dimer | HHblits | X-ray | 2.28Å | 0.27 | 0.38 | WSAF |
| 2f9f.1.A | 8.63 | monomer | HHblits | X-ray | 1.80Å | 0.26 | 0.38 | first mannosyl transferase (wbaZ-1) |
| 2x0f.1.A | 11.28 | homo-dimer | HHblits | X-ray | 2.55Å | 0.28 | 0.37 | WSAF |
| 5e9u.1.B | 7.69 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.36 | Glycosyltransferase-stabilizing protein Gtf2 |
| 5e9u.1.D | 7.69 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.36 | Glycosyltransferase-stabilizing protein Gtf2 |
| 5e9u.2.B | 7.69 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.36 | Glycosyltransferase-stabilizing protein Gtf2 |
| 5e9u.2.D | 7.69 | hetero-oligomer | HHblits | X-ray | 3.84Å | 0.26 | 0.36 | Glycosyltransferase-stabilizing protein Gtf2 |
| 4m83.1.A | 28.57 | monomer | BLAST | X-ray | 1.70Å | 0.35 | 0.31 | Oleandomycin glycosyltransferase |
| 4m83.2.A | 28.57 | monomer | BLAST | X-ray | 1.70Å | 0.35 | 0.31 | Oleandomycin glycosyltransferase |
| 4m60.1.A | 28.57 | monomer | BLAST | X-ray | 1.77Å | 0.35 | 0.31 | Oleandomycin glycosyltransferase |
| 2iyf.1.A | 28.57 | monomer | BLAST | X-ray | 1.70Å | 0.35 | 0.31 | OLEANDOMYCIN GLYCOSYLTRANSFERASE |
| 2iyf.2.A | 28.57 | monomer | BLAST | X-ray | 1.70Å | 0.35 | 0.31 | OLEANDOMYCIN GLYCOSYLTRANSFERASE |
| 1zxx.1.A | 13.91 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.26 | 0.32 | 6-phosphofructokinase |
| 6pfk.1.A | 12.17 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.26 | 0.32 | PHOSPHOFRUCTOKINASE |
| 3u39.1.A | 12.17 | homo-tetramer | HHblits | X-ray | 2.79Å | 0.26 | 0.32 | 6-phosphofructokinase |
| 4xyj.1.A | 9.73 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4xyk.1.A | 9.73 | homo-tetramer | HHblits | X-ray | 3.40Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4rh3.1.A | 9.73 | homo-tetramer | HHblits | X-ray | 3.02Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4u1r.1.B | 9.73 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4xz2.1.A | 8.85 | homo-tetramer | HHblits | X-ray | 2.67Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4xz2.1.B | 8.85 | homo-tetramer | HHblits | X-ray | 2.67Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4wl0.1.B | 8.85 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4wl0.1.C | 8.85 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4wl0.1.D | 8.85 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.27 | 0.31 | ATP-dependent 6-phosphofructokinase, platelet type |
| 3pfk.1.A | 12.28 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.26 | 0.31 | PHOSPHOFRUCTOKINASE |
| 4a3s.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.27 | 0.31 | 6-PHOSPHOFRUCTOKINASE |
| 2pfk.1.A | 19.82 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.31 | 6-PHOSPHOFRUCTOKINASE ISOZYME I |
| 1pfk.1.A | 19.82 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.31 | PHOSPHOFRUCTOKINASE |
| 4omt.1.A | 14.55 | homo-dimer | HHblits | X-ray | 6.00Å | 0.28 | 0.30 | 6-phosphofructokinase, muscle type |
| 3o8l.1.A | 14.55 | monomer | HHblits | X-ray | 3.20Å | 0.28 | 0.30 | 6-phosphofructokinase, muscle type |
| 3o8l.2.A | 14.55 | monomer | HHblits | X-ray | 3.20Å | 0.28 | 0.30 | 6-phosphofructokinase, muscle type |
| 3o8n.1.A | 14.55 | monomer | HHblits | X-ray | 3.20Å | 0.28 | 0.30 | 6-phosphofructokinase, muscle type |
| 3o8n.2.A | 14.55 | monomer | HHblits | X-ray | 3.20Å | 0.28 | 0.30 | 6-phosphofructokinase, muscle type |
| 4kq1.1.B | 13.64 | homo-tetramer | HHblits | X-ray | 2.66Å | 0.28 | 0.30 | Gsy2p |
| 4kq1.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.66Å | 0.28 | 0.30 | Gsy2p |
| 4kq1.1.C | 13.64 | homo-tetramer | HHblits | X-ray | 2.66Å | 0.28 | 0.30 | Gsy2p |
| 3rt1.1.A | 13.64 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.28 | 0.30 | PROTEIN (Glycogen [starch] synthase isoform 2) |
| 3rt1.1.B | 13.64 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.28 | 0.30 | PROTEIN (Glycogen [starch] synthase isoform 2) |
| 3rt1.1.C | 13.64 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.28 | 0.30 | PROTEIN (Glycogen [starch] synthase isoform 2) |
| 5sul.1.B | 13.64 | homo-tetramer | HHblits | X-ray | 3.30Å | 0.28 | 0.30 | Glycogen [starch] synthase isoform 2 |
| 5sul.1.C | 13.64 | homo-tetramer | HHblits | X-ray | 3.30Å | 0.28 | 0.30 | Glycogen [starch] synthase isoform 2 |
| 1mto.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 3.20Å | 0.26 | 0.31 | 6-phosphofructokinase |
| 3opy.1.A | 10.81 | hetero-oligomer | HHblits | X-ray | 3.05Å | 0.27 | 0.31 | 6-phosphofructo-1-kinase alpha-subunit |
| 3o8o.1.A | 10.81 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.27 | 0.31 | 6-phosphofructokinase subunit alpha |
| 4i4i.1.A | 14.41 | homo-tetramer | HHblits | X-ray | 2.49Å | 0.26 | 0.31 | 6-phosphofructokinase |
| 4i36.1.A | 14.41 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.31 | 6-phosphofructokinase |
| 4i7e.1.B | 14.41 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.26 | 0.31 | 6-phosphofructokinase |
| 4qlb.1.A | 12.84 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.30 | Probable glycogen [starch] synthase |
| 4qlb.1.B | 12.84 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.30 | Probable glycogen [starch] synthase |
| 5suk.1.A | 11.93 | homo-tetramer | HHblits | X-ray | 2.88Å | 0.27 | 0.30 | Glycogen [starch] synthase isoform 2 |
| 5suk.1.B | 11.93 | homo-tetramer | HHblits | X-ray | 2.88Å | 0.27 | 0.30 | Glycogen [starch] synthase isoform 2 |
| 5suk.1.C | 11.93 | homo-tetramer | HHblits | X-ray | 2.88Å | 0.27 | 0.30 | Glycogen [starch] synthase isoform 2 |
| 4kqm.1.B | 11.93 | homo-tetramer | HHblits | X-ray | 2.77Å | 0.27 | 0.30 | Gsy2p |
| 4kqm.1.A | 11.93 | homo-tetramer | HHblits | X-ray | 2.77Å | 0.27 | 0.30 | Gsy2p |
| 4kqm.1.C | 11.93 | homo-tetramer | HHblits | X-ray | 2.77Å | 0.27 | 0.30 | Gsy2p |
| 3rsz.1.A | 12.84 | homo-tetramer | HHblits | X-ray | 3.01Å | 0.27 | 0.30 | Glycogen [starch] synthase isoform 2 |
| 5jij.1.A | 13.33 | homo-tetramer | HHblits | X-ray | 1.82Å | 0.27 | 0.29 | Alpha,alpha-trehalose-phosphate synthase |
| 5jio.1.A | 13.33 | homo-tetramer | HHblits | X-ray | 1.71Å | 0.27 | 0.29 | Alpha,alpha-trehalose-phosphate synthase |
| 5l3k.2.D | 13.33 | homo-tetramer | HHblits | X-ray | 2.31Å | 0.27 | 0.29 | Alpha,alpha-trehalose-phosphate synthase |
| 5hvo.1.A | 15.24 | homo-dimer | HHblits | X-ray | 2.47Å | 0.27 | 0.29 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 3o8o.1.A | 18.63 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.28 | 0.28 | 6-phosphofructokinase subunit alpha |
| 3opy.1.A | 15.69 | hetero-oligomer | HHblits | X-ray | 3.05Å | 0.26 | 0.28 | 6-phosphofructo-1-kinase alpha-subunit |
| 1m6v.1.A | 23.96 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.31 | 0.27 | carbamoyl phosphate synthetase large chain |
| 3o8l.1.A | 15.69 | monomer | HHblits | X-ray | 3.20Å | 0.25 | 0.28 | 6-phosphofructokinase, muscle type |
| 3o8l.2.A | 15.69 | monomer | HHblits | X-ray | 3.20Å | 0.25 | 0.28 | 6-phosphofructokinase, muscle type |
| 3o8n.1.A | 15.69 | monomer | HHblits | X-ray | 3.20Å | 0.25 | 0.28 | 6-phosphofructokinase, muscle type |
| 3o8n.2.A | 15.69 | monomer | HHblits | X-ray | 3.20Å | 0.25 | 0.28 | 6-phosphofructokinase, muscle type |
| 4omt.1.A | 15.69 | homo-dimer | HHblits | X-ray | 6.00Å | 0.25 | 0.28 | 6-phosphofructokinase, muscle type |
| 1a9x.1.A | 19.59 | hetero-oligomer | HHblits | X-ray | 1.80Å | 0.29 | 0.27 | CARBAMOYL PHOSPHATE SYNTHETASE (LARGE CHAIN) |
| 5huv.1.A | 17.17 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.27 | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] |
| 5vz0.1.A | 13.00 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.26 | 0.28 | Pyruvate carboxylase |
| 5vz0.1.D | 13.00 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.26 | 0.28 | Pyruvate carboxylase |
| 5vyw.1.A | 12.00 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.25 | 0.28 | Pyruvate carboxylase |
| 5vyw.1.C | 12.00 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.25 | 0.28 | Pyruvate carboxylase |
| 1kzh.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.55Å | 0.28 | 0.27 | phosphofructokinase |
| 1kzh.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.55Å | 0.28 | 0.27 | phosphofructokinase |
| 2f48.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.11Å | 0.28 | 0.27 | diphosphate--fructose-6-phosphate 1-phosphotransferase |
| 4xyj.1.A | 15.00 | homo-tetramer | HHblits | X-ray | 3.10Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4xyk.1.A | 15.00 | homo-tetramer | HHblits | X-ray | 3.40Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4xz2.1.A | 15.00 | homo-tetramer | HHblits | X-ray | 2.67Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4xz2.1.B | 15.00 | homo-tetramer | HHblits | X-ray | 2.67Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4wl0.1.B | 15.00 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4wl0.1.C | 15.00 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4wl0.1.D | 15.00 | homo-tetramer | HHblits | X-ray | 2.89Å | 0.25 | 0.28 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4rap.1.A | 14.43 | homo-12-mer | HHblits | X-ray | 2.88Å | 0.27 | 0.27 | Glycosyltransferase TibC |
| 4rh3.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 3.02Å | 0.25 | 0.27 | ATP-dependent 6-phosphofructokinase, platelet type |
| 4u1r.1.B | 15.15 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.25 | 0.27 | ATP-dependent 6-phosphofructokinase, platelet type |
| 5v0t.1.A | 11.11 | monomer | HHblits | X-ray | 1.95Å | 0.25 | 0.27 | Alpha,alpha-trehalose-phosphate synthase (UDP-forming) |
| 3f5m.1.A | 9.80 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.23 | 0.28 | 6-phospho-1-fructokinase (ATP-dependent phosphofructokinase) |
| 2hig.1.A | 9.80 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.23 | 0.28 | 6-phospho-1-fructokinase |
| 5dou.1.A | 14.58 | homo-dimer | HHblits | X-ray | 2.60Å | 0.27 | 0.27 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
| 5dot.1.A | 14.58 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.27 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
| 4l8v.1.A | 21.35 | homo-tetramer | HHblits | X-ray | 2.09Å | 0.30 | 0.25 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 4mie.1.A | 19.78 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.25 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 4mie.1.D | 19.78 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.28 | 0.25 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 3ec7.1.A | 15.38 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.28 | 0.25 | Putative Dehydrogenase |
| 1cde.1.A | 22.35 | monomer | HHblits | X-ray | 2.50Å | 0.31 | 0.23 | PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE |
| 1gar.1.B | 22.35 | homo-dimer | HHblits | X-ray | 1.96Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1grc.1.A | 22.35 | homo-dimer | HHblits | X-ray | 3.00Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1grc.1.B | 22.35 | homo-dimer | HHblits | X-ray | 3.00Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1gar.1.A | 22.35 | homo-dimer | HHblits | X-ray | 1.96Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1c2t.1.A | 22.35 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1jkx.1.A | 22.35 | homo-dimer | HHblits | X-ray | 1.60Å | 0.31 | 0.23 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE |
| 1cdd.2.B | 22.35 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.23 | PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE |
| 1cdd.2.A | 22.35 | homo-dimer | HHblits | X-ray | 2.80Å | 0.31 | 0.23 | PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE |
| 1c3e.1.A | 22.35 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1ez1.1.A | 7.87 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.25 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE 2 |
| 1ez1.1.B | 7.87 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.25 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE 2 |
| 3gar.1.A | 22.35 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 2gar.1.A | 22.35 | monomer | HHblits | X-ray | 1.80Å | 0.31 | 0.23 | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE |
| 1kji.1.A | 7.95 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.24 | phosphoribosylglycinamide formyltransferase 2 |
| 1kji.1.B | 7.95 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.24 | phosphoribosylglycinamide formyltransferase 2 |
| 1rc0.1.A | 18.82 | monomer | HHblits | X-ray | 2.05Å | 0.30 | 0.23 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE |
| 1meo.1.A | 18.82 | monomer | HHblits | X-ray | 1.72Å | 0.30 | 0.23 | Phosphoribosylglycinamide formyltransferase |
| 2dwc.1.A | 13.64 | homo-dimer | HHblits | X-ray | 1.70Å | 0.27 | 0.24 | 433aa long hypothetical phosphoribosylglycinamide formyl transferase |
| 2czg.1.B | 13.64 | homo-dimer | HHblits | X-ray | 2.35Å | 0.27 | 0.24 | phosphoribosylglycinamide formyl transferase |
| 3da8.1.A | 19.05 | homo-dimer | HHblits | X-ray | 1.30Å | 0.31 | 0.23 | Probable 5'-phosphoribosylglycinamide formyltransferase purN |
| 3dcj.1.B | 19.05 | homo-dimer | HHblits | X-ray | 2.20Å | 0.31 | 0.23 | Probable 5'-phosphoribosylglycinamide formyltransferase purN |
| 4zyt.1.A | 19.05 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.23 | Trifunctional purine biosynthetic protein adenosine-3 |
| 4ds3.1.A | 15.29 | homo-dimer | HHblits | X-ray | 1.85Å | 0.28 | 0.23 | Phosphoribosylglycinamide formyltransferase |
| 3p9x.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.23 | phosphoribosylglycinamide formyltransferase |
| 3av3.1.A | 16.47 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.23 | Phosphoribosylglycinamide formyltransferase |
| 3w7b.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.71Å | 0.28 | 0.23 | Formyltetrahydrofolate deformylase |
| 3o1l.1.A | 8.33 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.23 | Formyltetrahydrofolate deformylase |
| 2ywr.1.A | 15.48 | monomer | HHblits | X-ray | 1.77Å | 0.27 | 0.23 | Phosphoribosylglycinamide formyltransferase |
| 5cjj.1.A | 8.24 | homo-dimer | HHblits | X-ray | 2.42Å | 0.26 | 0.23 | Phosphoribosylglycinamide formyltransferase |
| 5cjj.1.B | 8.24 | homo-dimer | HHblits | X-ray | 2.42Å | 0.26 | 0.23 | Phosphoribosylglycinamide formyltransferase |
| 3lou.1.A | 13.10 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.23 | Formyltetrahydrofolate deformylase |
| 3lou.1.B | 13.10 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.27 | 0.23 | Formyltetrahydrofolate deformylase |
| 3n0v.1.A | 9.52 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.26 | 0.23 | Formyltetrahydrofolate deformylase |
| 4e2f.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1r0b.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1za2.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1za2.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1za1.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1za1.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 3d7s.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 3d7s.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 3csu.1.A | 17.07 | homo-trimer | HHblits | X-ray | 1.88Å | 0.28 | 0.23 | PROTEIN (ASPARTATE CARBAMOYLTRANSFERASE) |
| 3csu.1.B | 17.07 | homo-trimer | HHblits | X-ray | 1.88Å | 0.28 | 0.23 | PROTEIN (ASPARTATE CARBAMOYLTRANSFERASE) |
| 2qg9.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 2qg9.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 2qgf.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 2qgf.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.20Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 2air.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 4fyv.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 4fyv.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 2fzg.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.25Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 2fzg.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.25Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1rac.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1raf.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1acm.1.A | 18.29 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.23 | ASPARTATE CARBAMOYLTRANSFERASE, CATALYTIC CHAIN |
| 1sku.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1sku.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.28 | 0.23 | Aspartate carbamoyltransferase catalytic chain |
| 1i5o.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.23 | ASPARTATE TRANSCARBAMOYLASE CATALYTIC CHAIN |
| 1i5o.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.28 | 0.23 | ASPARTATE TRANSCARBAMOYLASE CATALYTIC CHAIN |
| 5vmq.1.B | 17.07 | homo-trimer | HHblits | X-ray | 2.01Å | 0.28 | 0.23 | Aspartate carbamoyltransferase |
| 5vmq.1.C | 17.07 | homo-trimer | HHblits | X-ray | 2.01Å | 0.28 | 0.23 | Aspartate carbamoyltransferase |
| 5vmq.1.A | 17.07 | homo-trimer | HHblits | X-ray | 2.01Å | 0.28 | 0.23 | Aspartate carbamoyltransferase |
| 1f1b.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.23 | ASPARTATE CARBAMOYLTRANSFERASE CATALYTIC CHAIN |
| 1ezz.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.28 | 0.23 | ASPARTATE CARBAMOYLTRANSFERASE CATALYTIC CHAIN |
| 1ezz.1.C | 17.07 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.28 | 0.23 | ASPARTATE CARBAMOYLTRANSFERASE CATALYTIC CHAIN |
| 3auf.1.A | 14.46 | homo-dimer | HHblits | X-ray | 2.07Å | 0.27 | 0.23 | Glycinamide ribonucleotide transformylase 1 |
| 9atc.1.A | 17.07 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.27 | 0.23 | ASPARTATE TRANSCARBAMOYLASE |
| 4zj8.1.A | 10.98 | monomer | HHblits | X-ray | 2.75Å | 0.27 | 0.23 | human OX1R fusion protein to P.abysii glycogen synthase |
| 4npa.1.A | 16.46 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.28 | 0.22 | Putative uncharacterized protein |
| 3kcq.1.A | 17.95 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.22 | Phosphoribosylglycinamide formyltransferase |
| 3kcq.1.B | 17.95 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.22 | Phosphoribosylglycinamide formyltransferase |
| 3gh1.1.A | 15.19 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.28 | 0.22 | Predicted nucleotide-binding protein |
| 3gh1.1.B | 15.19 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.28 | 0.22 | Predicted nucleotide-binding protein |
| 3ngl.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.18 | Bifunctional protein folD |
| 4zmr.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.17 | Response regulator |
| 4zmr.1.B | 15.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.17 | Response regulator |
| 2qip.1.A | 12.07 | homo-dimer | HHblits | X-ray | 1.48Å | 0.28 | 0.16 | Protein of unknown function VPA0982 |
| 3afo.1.A | 14.04 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.16 | NADH kinase POS5 |
| 3afo.1.B | 14.04 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.16 | NADH kinase POS5 |
| 4ldz.1.A | 17.24 | homo-dimer | HHblits | X-ray | 2.31Å | 0.27 | 0.16 | Transcriptional regulatory protein DesR |
| 4ldz.1.B | 17.24 | homo-dimer | HHblits | X-ray | 2.31Å | 0.27 | 0.16 | Transcriptional regulatory protein DesR |
| 3pfn.1.A | 7.02 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.28 | 0.16 | NAD kinase |
| 3io5.1.A | 16.98 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.15 | Recombination and repair protein |
| 1yt5.1.B | 15.38 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.14 | inorganic polyphosphate/ATP-NAD kinase |
| 1yt5.1.A | 15.38 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.14 | inorganic polyphosphate/ATP-NAD kinase |
| 1yt5.2.B | 15.38 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.14 | inorganic polyphosphate/ATP-NAD kinase |
| 2qip.1.A | 16.00 | homo-dimer | HHblits | X-ray | 1.48Å | 0.27 | 0.14 | Protein of unknown function VPA0982 |
| 1dia.1.A | 12.77 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.13 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE |
| 1dia.1.B | 12.77 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.13 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE |
| 3q7s.1.A | 14.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.13 | Transcriptional regulatory protein |
| 3q7s.1.B | 14.89 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.13 | Transcriptional regulatory protein |
| 3q7t.1.A | 14.89 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.13 | Transcriptional regulatory protein |
| 3q7t.1.B | 14.89 | homo-dimer | HHblits | X-ray | 2.15Å | 0.26 | 0.13 | Transcriptional regulatory protein |
| 3jzd.1.A | 17.78 | homo-dimer | HHblits | X-ray | 2.10Å | 0.29 | 0.12 | Iron-containing alcohol dehydrogenase |
| 2ahd.1.A | 27.91 | monomer | HHblits | X-ray | 3.00Å | 0.32 | 0.12 | Phosphodiesterase MJ0936 |
| 1s3m.1.A | 27.91 | monomer | HHblits | X-ray | 2.50Å | 0.32 | 0.12 | Hypothetical protein MJ0936 |
| 1s3l.1.A | 27.91 | monomer | HHblits | X-ray | 2.40Å | 0.32 | 0.12 | Hypothetical protein MJ0936 |
| 4x7g.1.A | 18.60 | monomer | HHblits | X-ray | 1.22Å | 0.29 | 0.12 | Precorrin-6A reductase |
| 5c4n.1.A | 18.60 | monomer | HHblits | X-ray | 1.63Å | 0.29 | 0.12 | Precorrin-6A reductase |
| 5c4r.1.A | 18.60 | monomer | HHblits | X-ray | 3.17Å | 0.29 | 0.12 | Precorrin-6A reductase |
| 5h98.1.A | 16.28 | monomer | HHblits | X-ray | 2.04Å | 0.29 | 0.12 | Geobacter metallireducens SMUG1 |
| 5h99.1.A | 16.28 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.12 | Geobacter metallireducens SMUG1 |
| 5h93.1.A | 16.28 | monomer | HHblits | X-ray | 2.18Å | 0.29 | 0.12 | Geobacter metallireducens SMUG1 |
| 5h9i.1.A | 16.28 | monomer | HHblits | X-ray | 1.50Å | 0.29 | 0.12 | Geobacter metallireducens SMUG1 |
| 5c4r.1.A | 21.95 | monomer | HHblits | X-ray | 3.17Å | 0.32 | 0.11 | Precorrin-6A reductase |
| 4x7g.1.A | 21.95 | monomer | HHblits | X-ray | 1.22Å | 0.32 | 0.11 | Precorrin-6A reductase |
| 5c4n.1.A | 21.95 | monomer | HHblits | X-ray | 1.63Å | 0.32 | 0.11 | Precorrin-6A reductase |
| 1u6k.1.A | 16.67 | homo-hexamer | HHblits | X-ray | 1.55Å | 0.30 | 0.12 | F420-dependent methylenetetrahydromethanopterin dehydrogenase |
| 1u6k.1.A | 24.39 | homo-hexamer | HHblits | X-ray | 1.55Å | 0.32 | 0.11 | F420-dependent methylenetetrahydromethanopterin dehydrogenase |
| 1oe4.1.A | 13.95 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.12 | SINGLE-STRAND SELECTIVE MONOFUNCTIONAL URACIL DNA GLYCOSYLASE |
| 1oe5.1.B | 13.95 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.12 | SINGLE-STRAND SELECTIVE MONOFUNCTIONAL URACIL DNA GLYCOSYLASE |
| 3bil.1.A | 14.63 | homo-dimer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | Probable LacI-family transcriptional regulator |
| 1z2w.1.A | 14.29 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.12 | Vacuolar protein sorting 29 |
| 1z2w.2.A | 14.29 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.12 | Vacuolar protein sorting 29 |
| 3pso.1.A | 14.29 | monomer | HHblits | X-ray | 3.20Å | 0.27 | 0.12 | Vacuolar protein sorting-associated protein 29 |
| 1w24.1.A | 14.29 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.12 | VACUOLAR PROTEIN SORTING PROTEIN 29 |
| 2r17.1.A | 14.29 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.27 | 0.12 | Vacuolar protein sorting-associated protein 29 |
| 3k4h.1.A | 9.52 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.12 | putative transcriptional regulator |
| 3k4h.1.B | 9.52 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.12 | putative transcriptional regulator |
| 3h5t.1.A | 17.50 | monomer | HHblits | X-ray | 2.53Å | 0.30 | 0.11 | Transcriptional regulator, LacI family |
| 3oqo.1.A | 15.00 | hetero-oligomer | HHblits | X-ray | 2.97Å | 0.30 | 0.11 | Catabolite control protein A |
| 3oqo.1.C | 15.00 | hetero-oligomer | HHblits | X-ray | 2.97Å | 0.30 | 0.11 | Catabolite control protein A |
| 3oqn.1.A | 15.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.11 | Catabolite control protein A |
| 3oqn.1.C | 15.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.11 | Catabolite control protein A |
| 3huu.1.A | 12.20 | homo-dimer | HHblits | X-ray | 1.95Å | 0.28 | 0.11 | Transcription regulator like protein |
| 2a22.1.A | 17.07 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.11 | vacuolar protein sorting 29 |
| 2a22.2.A | 17.07 | monomer | HHblits | X-ray | 2.20Å | 0.28 | 0.11 | vacuolar protein sorting 29 |
| 1rzr.1.C | 20.00 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1rzr.1.D | 20.00 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1rzr.2.C | 20.00 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1rzr.2.D | 20.00 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 2hsg.1.A | 20.00 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 5iun.3.D | 19.51 | hetero-oligomer | HHblits | X-ray | 2.79Å | 0.27 | 0.11 | Transcriptional regulatory protein DesR |
| 4le1.2.A | 19.51 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.11 | Transcriptional regulatory protein DesR |
| 4le2.1.A | 19.51 | homo-dimer | HHblits | X-ray | 2.54Å | 0.27 | 0.11 | Transcriptional regulatory protein DesR |
| 4le0.1.A | 19.51 | homo-dimer | HHblits | X-ray | 2.27Å | 0.27 | 0.11 | Transcriptional regulatory protein DesR |
| 4le1.1.A | 19.51 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.11 | Transcriptional regulatory protein DesR |
| 5iun.1.C | 19.51 | hetero-oligomer | HHblits | X-ray | 2.79Å | 0.27 | 0.11 | Transcriptional regulatory protein DesR |
| 2fep.1.A | 15.38 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.30 | 0.11 | Catabolite control protein A |
| 3ikt.1.A | 11.90 | homo-dimer | HHblits | X-ray | 2.26Å | 0.24 | 0.12 | Redox-sensing transcriptional repressor rex |
| 3ikv.1.A | 11.90 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.12 | Redox-sensing transcriptional repressor rex |
| 3ikv.1.B | 11.90 | homo-dimer | HHblits | X-ray | 2.40Å | 0.24 | 0.12 | Redox-sensing transcriptional repressor rex |
| 3il2.1.A | 11.90 | homo-dimer | HHblits | X-ray | 2.49Å | 0.24 | 0.12 | Redox-sensing transcriptional repressor rex |
| 3il2.1.B | 11.90 | homo-dimer | HHblits | X-ray | 2.49Å | 0.24 | 0.12 | Redox-sensing transcriptional repressor rex |
| 1c30.1.A | 27.03 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.34 | 0.10 | CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT |
| 1bxr.1.A | 27.03 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.34 | 0.10 | CARBAMOYL-PHOSPHATE SYNTHASE |
| 3hcw.1.A | 12.20 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.11 | Maltose operon transcriptional repressor |
| 3hcw.1.B | 12.20 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.11 | Maltose operon transcriptional repressor |
| 4la4.1.A | 26.32 | homo-tetramer | HHblits | X-ray | 2.07Å | 0.32 | 0.10 | NAD(P)H dehydrogenase (quinone) |
| 4laf.1.D | 26.32 | homo-tetramer | HHblits | X-ray | 1.76Å | 0.32 | 0.10 | NAD(P)H dehydrogenase (quinone) |
| 2oen.1.A | 20.51 | hetero-oligomer | HHblits | X-ray | 3.17Å | 0.29 | 0.11 | Catabolite control protein |
| 1sxi.1.B | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.2.A | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.2.B | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.3.A | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.3.B | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.4.A | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.4.B | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.5.A | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.5.B | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.6.A | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.6.B | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxh.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxh.1.B | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxg.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxg.1.B | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxg.2.A | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxg.2.B | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxg.3.A | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxg.3.B | 20.51 | homo-dimer | HHblits | X-ray | 2.75Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1sxi.1.A | 20.51 | homo-dimer | HHblits | X-ray | 3.00Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 2nzu.1.A | 20.51 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.29 | 0.11 | Catabolite control protein |
| 2jcg.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.11 | GLUCOSE-RESISTANCE AMYLASE REGULATOR |
| 1zvv.2.B | 20.51 | hetero-oligomer | HHblits | X-ray | 2.98Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1zvv.3.B | 20.51 | hetero-oligomer | HHblits | X-ray | 2.98Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 1zvv.1.B | 20.51 | hetero-oligomer | HHblits | X-ray | 2.98Å | 0.29 | 0.11 | Glucose-resistance amylase regulator |
| 3kjx.1.B | 23.08 | homo-dimer | HHblits | X-ray | 2.33Å | 0.29 | 0.11 | Transcriptional regulator, LacI family |
| 3kjx.1.A | 23.08 | homo-dimer | HHblits | X-ray | 2.33Å | 0.29 | 0.11 | Transcriptional regulator, LacI family |
| 3kjx.2.B | 23.08 | homo-dimer | HHblits | X-ray | 2.33Å | 0.29 | 0.11 | Transcriptional regulator, LacI family |
| 2ynm.1.D | 22.22 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.36 | 0.10 | LIGHT-INDEPENDENT PROTOCHLOROPHYLLIDE REDUCTASE SUBUNIT B |
| 5xcj.1.A | 9.52 | monomer | HHblits | X-ray | 1.86Å | 0.23 | 0.12 | Vacuolar protein sorting-associated protein 29 |
| 5xcj.2.A | 9.52 | monomer | HHblits | X-ray | 1.86Å | 0.23 | 0.12 | Vacuolar protein sorting-associated protein 29 |
| 5xck.1.A | 9.52 | monomer | HHblits | X-ray | 2.20Å | 0.23 | 0.12 | Vacuolar protein sorting-associated protein 29 |
| 5xck.2.A | 9.52 | monomer | HHblits | X-ray | 2.20Å | 0.23 | 0.12 | Vacuolar protein sorting-associated protein 29 |
| 3ezy.1.A | 17.95 | homo-tetramer | HHblits | X-ray | 2.04Å | 0.29 | 0.11 | Dehydrogenase |
| 4rk1.1.A | 28.95 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.10 | Ribose transcriptional regulator |
| 1jhz.1.A | 7.50 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.11 | PURINE NUCLEOTIDE SYNTHESIS REPRESSOR |
| 1jhz.1.B | 7.50 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.11 | PURINE NUCLEOTIDE SYNTHESIS REPRESSOR |
| 4l9r.1.A | 23.08 | homo-tetramer | HHblits | X-ray | 1.95Å | 0.29 | 0.11 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 3kke.1.B | 17.95 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.11 | LacI family Transcriptional regulator |
| 3kke.1.A | 17.95 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.11 | LacI family Transcriptional regulator |
| 3gv0.1.A | 20.51 | monomer | HHblits | X-ray | 2.35Å | 0.28 | 0.11 | Transcriptional regulator, LacI family |
| 5ax7.1.A | 24.32 | homo-dimer | HHblits | X-ray | 2.46Å | 0.32 | 0.10 | Pyruvyl transferase 1 |
| 3aer.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.34 | 0.10 | Light-independent protochlorophyllide reductase subunit B |
| 3aeq.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.34 | 0.10 | Light-independent protochlorophyllide reductase subunit B |
| 3aek.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.34 | 0.10 | Light-independent protochlorophyllide reductase subunit B |
| 3aeu.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.34 | 0.10 | Light-independent protochlorophyllide reductase subunit B |
| 3aet.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.91Å | 0.34 | 0.10 | Light-independent protochlorophyllide reductase subunit B |
| 3qk7.1.A | 10.00 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.11 | Transcriptional regulators |
| 3qk7.2.A | 10.00 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.11 | Transcriptional regulators |
| 3qk7.2.B | 10.00 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.11 | Transcriptional regulators |
| 3bbl.1.A | 10.00 | homo-dimer | HHblits | X-ray | 2.35Å | 0.26 | 0.11 | Regulatory protein of LacI family |
| 3vrd.1.B | 35.29 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.39 | 0.09 | Flavocytochrome c flavin subunit |
| 3evn.1.A | 20.51 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.11 | Oxidoreductase, Gfo/Idh/MocA family |
| 5dou.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.60Å | 0.34 | 0.10 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
| 5dot.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.40Å | 0.34 | 0.10 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
| 5b51.1.A | 18.42 | monomer | HHblits | X-ray | 1.30Å | 0.30 | 0.10 | ABC-type transporter, periplasmic component |
| 5az3.1.A | 18.42 | monomer | HHblits | X-ray | 1.42Å | 0.30 | 0.10 | ABC-type transporter, periplasmic component |
| 5b50.1.A | 18.42 | monomer | HHblits | X-ray | 1.65Å | 0.30 | 0.10 | ABC-type transporter, periplasmic component |
| 5b4z.1.A | 18.42 | monomer | HHblits | X-ray | 1.30Å | 0.30 | 0.10 | ABC-type transporter, periplasmic component |
| 1vdm.1.B | 21.05 | homo-12-mer | HHblits | X-ray | 2.50Å | 0.30 | 0.10 | purine phosphoribosyltransferase |
| 1vdm.1.A | 21.05 | homo-12-mer | HHblits | X-ray | 2.50Å | 0.30 | 0.10 | purine phosphoribosyltransferase |
| 1vdm.1.E | 21.05 | homo-12-mer | HHblits | X-ray | 2.50Å | 0.30 | 0.10 | purine phosphoribosyltransferase |
| 1c30.1.A | 18.92 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.32 | 0.10 | CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT |
| 1bxr.1.A | 18.92 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.32 | 0.10 | CARBAMOYL-PHOSPHATE SYNTHASE |
| 3wgi.1.B | 15.00 | homo-dimer | HHblits | X-ray | 3.25Å | 0.25 | 0.11 | Redox-sensing transcriptional repressor rex |
| 3wg9.2.A | 15.00 | homo-dimer | HHblits | X-ray | 1.97Å | 0.25 | 0.11 | Redox-sensing transcriptional repressor rex |
| 3wg9.2.B | 15.00 | homo-dimer | HHblits | X-ray | 1.97Å | 0.25 | 0.11 | Redox-sensing transcriptional repressor rex |
| 3wgh.1.B | 15.00 | homo-dimer | HHblits | X-ray | 2.05Å | 0.25 | 0.11 | Redox-sensing transcriptional repressor rex |
| 3wgg.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.11 | Redox-sensing transcriptional repressor rex |
| 3wgg.1.B | 15.00 | homo-dimer | HHblits | X-ray | 2.10Å | 0.25 | 0.11 | Redox-sensing transcriptional repressor rex |
| 1dbq.1.A | 7.69 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | PURINE REPRESSOR |
| 1dbq.1.B | 7.69 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.11 | PURINE REPRESSOR |
| 4rk6.1.A | 12.20 | homo-dimer | HHblits | X-ray | 1.76Å | 0.23 | 0.11 | Glucose-resistance amylase regulator |
| 1fcd.1.A | 36.36 | hetero-oligomer | HHblits | X-ray | 2.53Å | 0.40 | 0.09 | FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT) |
| 1fcd.2.A | 36.36 | hetero-oligomer | HHblits | X-ray | 2.53Å | 0.40 | 0.09 | FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT) |
| 2o20.1.A | 10.26 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.11 | Catabolite control protein A |
| 2o20.1.B | 10.26 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.11 | Catabolite control protein A |
| 3e3m.1.A | 7.69 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.11 | Transcriptional regulator, LacI family |
| 4rb4.1.A | 10.26 | homo-12-mer | HHblits | X-ray | 3.88Å | 0.26 | 0.11 | Glycosyltransferase tibC |
| 2q5c.1.A | 17.95 | monomer | HHblits | X-ray | 1.49Å | 0.26 | 0.11 | NtrC family transcriptional regulator |
| 3brq.1.A | 10.26 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.11 | HTH-type transcriptional regulator ascG |
| 3brq.1.B | 10.26 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.11 | HTH-type transcriptional regulator ascG |
| 4koa.1.A | 18.42 | homo-dimer | HHblits | X-ray | 1.93Å | 0.28 | 0.10 | 1,5-anhydro-D-fructose reductase |
| 5m29.1.A | 21.62 | monomer | HHblits | X-ray | 1.50Å | 0.30 | 0.10 | Vitamin B12-binding protein |
| 5m2q.1.A | 21.62 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.10 | Outer membrane protein A,Vitamin B12-binding protein |
| 5m34.1.A | 21.62 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.10 | Vitamin B12-binding protein |
| 5m34.2.A | 21.62 | monomer | HHblits | X-ray | 1.60Å | 0.30 | 0.10 | Vitamin B12-binding protein |
| 3h5o.1.A | 18.42 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.10 | Transcriptional regulator GntR |
| 3r5f.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.07Å | 0.30 | 0.10 | D-alanine--D-alanine ligase 1 |
| 1a9x.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 1.80Å | 0.32 | 0.10 | CARBAMOYL PHOSPHATE SYNTHETASE (LARGE CHAIN) |
| 1m6v.1.A | 19.44 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.32 | 0.10 | carbamoyl phosphate synthetase large chain |
| 1ydh.1.A | 10.53 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.10 | At5g11950 |
| 2q4d.1.B | 10.53 | homo-dimer | HHblits | X-ray | 2.15Å | 0.27 | 0.10 | Lysine decarboxylase-like protein At5g11950 |
| 3e5n.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.10 | D-alanine-D-alanine ligase A |
| 3cwc.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.23Å | 0.31 | 0.10 | Putative glycerate kinase 2 |
| 3cwc.1.B | 16.67 | homo-dimer | HHblits | X-ray | 2.23Å | 0.31 | 0.10 | Putative glycerate kinase 2 |
| 2a33.1.A | 22.22 | homo-dimer | HHblits | X-ray | 1.95Å | 0.31 | 0.10 | hypothetical protein |
| 2q4o.1.B | 22.22 | homo-dimer | HHblits | X-ray | 1.95Å | 0.31 | 0.10 | Uncharacterized protein At2g37210/T2N18.3 |
| 4m7o.1.A | 16.67 | monomer | HHblits | X-ray | 2.02Å | 0.31 | 0.10 | Iron-binding protein |
| 3l49.1.A | 21.62 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.10 | ABC sugar (Ribose) transporter, periplasmic substrate-binding subunit |
| 1urp.1.A | 21.62 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.10 | D-RIBOSE-BINDING PROTEIN |
| 2dri.1.A | 21.62 | monomer | HHblits | X-ray | 1.60Å | 0.29 | 0.10 | D-RIBOSE-BINDING PROTEIN |
| 1drj.1.A | 21.62 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.10 | D-RIBOSE-BINDING PROTEIN |
| 4rie.1.A | 31.43 | homo-dimer | HHblits | X-ray | 2.16Å | 0.33 | 0.10 | Glycosyl transferase homolog |
| 4rie.1.B | 31.43 | homo-dimer | HHblits | X-ray | 2.16Å | 0.33 | 0.10 | Glycosyl transferase homolog |
| 4rif.1.A | 31.43 | homo-dimer | HHblits | X-ray | 1.85Å | 0.33 | 0.10 | Glycosyl transferase homolog |
| 4rif.1.B | 31.43 | homo-dimer | HHblits | X-ray | 1.85Å | 0.33 | 0.10 | Glycosyl transferase homolog |
| 5gj3.1.A | 19.44 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.10 | Periplasmic binding protein |
| 5y8b.1.A | 19.44 | monomer | HHblits | X-ray | 2.40Å | 0.31 | 0.10 | Periplasmic binding protein |
| 2gmh.1.A | 13.51 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.10 | Electron transfer flavoprotein-ubiquinone oxidoreductase |
| 1l5x.1.A | 25.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.10 | Survival protein E |
| 1l5x.1.B | 25.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.10 | Survival protein E |
| 4rap.1.A | 5.13 | homo-12-mer | HHblits | X-ray | 2.88Å | 0.24 | 0.11 | Glycosyltransferase TibC |
| 1l1q.1.A | 13.16 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.10 | Adenine phosphoribosyltransferase |
| 2rgy.1.A | 10.26 | homo-dimer | HHblits | X-ray | 2.05Å | 0.24 | 0.11 | Transcriptional regulator, LacI family |
| 3gd3.1.A | 26.47 | monomer | HHblits | X-ray | 2.95Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.2.A | 26.47 | monomer | HHblits | X-ray | 2.95Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.4.A | 26.47 | monomer | HHblits | X-ray | 2.95Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 5fs8.1.A | 26.47 | monomer | HHblits | X-ray | 1.40Å | 0.35 | 0.09 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 1gv4.1.A | 26.47 | monomer | HHblits | X-ray | 2.00Å | 0.35 | 0.09 | PROGRAMED CELL DEATH PROTEIN 8 |
| 5fs6.1.A | 26.47 | monomer | HHblits | X-ray | 1.90Å | 0.35 | 0.09 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5miu.1.A | 26.47 | monomer | HHblits | X-ray | 3.50Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.A | 26.47 | homo-dimer | HHblits | X-ray | 3.10Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.B | 26.47 | homo-dimer | HHblits | X-ray | 3.10Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 1ker.1.A | 29.41 | homo-dimer | HHblits | X-ray | 2.20Å | 0.35 | 0.09 | dTDP-D-glucose 4,6-dehydratase |
| 4lza.1.A | 13.16 | homo-dimer | HHblits | X-ray | 1.84Å | 0.26 | 0.10 | Adenine phosphoribosyltransferase |
| 1n4a.2.A | 22.22 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.10 | Vitamin B12 transport protein btuF |
| 1n4d.1.A | 22.22 | monomer | HHblits | X-ray | 3.00Å | 0.30 | 0.10 | Vitamin B12 transport protein btuF |
| 1n4d.2.A | 22.22 | monomer | HHblits | X-ray | 3.00Å | 0.30 | 0.10 | Vitamin B12 transport protein btuF |
| 1n2z.3.A | 22.22 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.10 | Vitamin B12 transport protein btuF |
| 2qi9.1.E | 22.22 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.10 | Vitamin B12-binding protein btuF |
| 1n2z.3.B | 22.22 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.10 | Vitamin B12 transport protein btuF |
| 5m3b.1.A | 22.22 | monomer | HHblits | X-ray | 1.50Å | 0.30 | 0.10 | Vitamin B12-binding protein |
| 5khl.1.A | 16.22 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.10 | Hemin ABC transporter, periplasmic hemin-binding protein HutB |
| 5b6h.1.A | 15.79 | homo-dimer | HHblits | X-ray | 1.90Å | 0.26 | 0.10 | Adenine phosphoribosyltransferase |
| 4mb6.1.A | 15.79 | homo-dimer | HHblits | X-ray | 1.81Å | 0.26 | 0.10 | Adenine phosphoribosyltransferase |
| 3bq9.1.A | 25.71 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.32 | 0.10 | Predicted Rossmann fold nucleotide-binding domain-containing protein |
| 5cr9.1.A | 20.00 | monomer | HHblits | X-ray | 1.70Å | 0.32 | 0.10 | ABC-type Fe3+-hydroxamate transport system, periplasmic component |
| 2xdq.1.B | 13.89 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.30 | 0.10 | LIGHT-INDEPENDENT PROTOCHLOROPHYLLIDE REDUCTASE SUBUNIT B |
| 3nt5.1.A | 26.47 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.34 | 0.09 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase |
| 5b58.1.E | 14.29 | hetero-oligomer | HHblits | X-ray | 3.21Å | 0.32 | 0.10 | Putative hemin transport system, substrate-binding protein |
| 5giz.2.A | 14.29 | monomer | HHblits | X-ray | 1.50Å | 0.32 | 0.10 | Putative hemin transport system, substrate-binding protein |
| 5y89.2.A | 14.29 | monomer | HHblits | X-ray | 2.40Å | 0.32 | 0.10 | Putative hemin transport system, substrate-binding protein |
| 5y8a.2.A | 14.29 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.10 | Putative hemin transport system, substrate-binding protein |
| 5y8a.1.A | 14.29 | monomer | HHblits | X-ray | 2.00Å | 0.32 | 0.10 | Putative hemin transport system, substrate-binding protein |
| 3egc.1.A | 7.69 | homo-dimer | HHblits | X-ray | 2.35Å | 0.23 | 0.11 | putative ribose operon repressor |
| 4fdc.1.A | 23.53 | monomer | HHblits | X-ray | 2.40Å | 0.34 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 5fs7.1.A | 23.53 | monomer | HHblits | X-ray | 1.85Å | 0.34 | 0.09 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs9.1.A | 23.53 | monomer | HHblits | X-ray | 1.75Å | 0.34 | 0.09 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 3gd4.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.24Å | 0.34 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd4.1.B | 23.53 | homo-dimer | HHblits | X-ray | 2.24Å | 0.34 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 5fmh.1.A | 23.53 | monomer | HHblits | X-ray | 1.80Å | 0.34 | 0.09 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 2khz.1.A | 20.00 | homo-dimer | HHblits | NMR | NA | 0.31 | 0.10 | c-Myc-responsive protein Rcl |
| 4ru1.1.A | 16.22 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.10 | Monosaccharide ABC transporter substrate-binding protein, CUT2 family |
| 4dbl.1.E | 22.86 | hetero-oligomer | HHblits | X-ray | 3.49Å | 0.31 | 0.10 | Vitamin B12-binding protein |
| 4fi3.1.E | 22.86 | hetero-oligomer | HHblits | X-ray | 3.47Å | 0.31 | 0.10 | Vitamin B12-binding protein |
| 4m0k.1.A | 7.69 | homo-dimer | HHblits | X-ray | 1.40Å | 0.22 | 0.11 | Adenine phosphoribosyltransferase |
| 4m0k.2.B | 7.69 | homo-dimer | HHblits | X-ray | 1.40Å | 0.22 | 0.11 | Adenine phosphoribosyltransferase |
| 2gx6.1.A | 19.44 | monomer | HHblits | X-ray | 1.97Å | 0.28 | 0.10 | D-ribose-binding periplasmic protein |
| 5kvi.1.A | 27.27 | monomer | HHblits | X-ray | 2.00Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 1m6i.1.A | 27.27 | monomer | HHblits | X-ray | 1.80Å | 0.35 | 0.09 | Programmed cell death protein 8 |
| 5kvh.1.A | 27.27 | homo-dimer | HHblits | X-ray | 2.27Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 3d8u.1.A | 13.51 | homo-dimer | HHblits | X-ray | 2.88Å | 0.26 | 0.10 | PurR transcriptional regulator |
| 3d8u.1.B | 13.51 | homo-dimer | HHblits | X-ray | 2.88Å | 0.26 | 0.10 | PurR transcriptional regulator |
| 4bur.1.A | 24.24 | monomer | HHblits | X-ray | 2.88Å | 0.35 | 0.09 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 4bur.3.A | 24.24 | monomer | HHblits | X-ray | 2.88Å | 0.35 | 0.09 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 4lii.1.A | 24.24 | monomer | HHblits | X-ray | 1.88Å | 0.35 | 0.09 | Apoptosis-inducing factor 1, mitochondrial |
| 2r79.1.A | 22.86 | homo-hexamer | HHblits | X-ray | 2.40Å | 0.30 | 0.10 | Periplasmic binding protein |
| 3o75.1.A | 10.81 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.10 | Fructose transport system repressor FruR |
| 3o74.1.B | 10.81 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.10 | Fructose transport system repressor FruR |
| 3hyv.1.A | 24.24 | homo-trimer | HHblits | X-ray | 2.30Å | 0.34 | 0.09 | Sulfide-quinone reductase |
| 3pdi.1.B | 21.21 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.34 | 0.09 | Nitrogenase MoFe cofactor biosynthesis protein NifN |
| 2bc1.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.15Å | 0.32 | 0.09 | NADH Oxidase |
| 2bc0.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.00Å | 0.32 | 0.09 | NADH Oxidase |
| 3ics.1.A | 26.47 | homo-dimer | HHblits | X-ray | 1.94Å | 0.32 | 0.09 | Coenzyme A-Disulfide Reductase |
| 4toi.1.A | 23.53 | homo-dimer | HHblits | X-ray | 2.30Å | 0.32 | 0.09 | 30S ribosomal protein S2,Ribosomal protein S1 |
| 3md9.1.A | 8.33 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.10 | Hemin-binding periplasmic protein hmuT |
| 3md9.2.A | 8.33 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.10 | Hemin-binding periplasmic protein hmuT |
| 2dy0.1.A | 13.51 | homo-dimer | HHblits | X-ray | 1.25Å | 0.25 | 0.10 | Adenine phosphoribosyltransferase |
| 2rg7.1.B | 13.89 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.10 | Periplasmic HEME binding protein |
| 2rg7.1.A | 13.89 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.10 | Periplasmic HEME binding protein |
| 2r7a.2.B | 13.89 | homo-dimer | HHblits | X-ray | 2.05Å | 0.26 | 0.10 | Bacterial heme binding protein |
| 1drk.1.A | 20.00 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.10 | D-RIBOSE-BINDING PROTEIN |
| 4eqx.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.70Å | 0.30 | 0.09 | Coenzyme A disulfide reductase |
| 4em3.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.98Å | 0.30 | 0.09 | Coenzyme A disulfide reductase |
| 5lbt.1.A | 8.11 | homo-dimer | HHblits | X-ray | 1.75Å | 0.24 | 0.10 | Ribosyldihydronicotinamide dehydrogenase [quinone] |
| 3o73.1.A | 8.11 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.10 | Ribosyldihydronicotinamide dehydrogenase [quinone] |
| 3o73.1.B | 8.11 | homo-dimer | HHblits | X-ray | 2.00Å | 0.24 | 0.10 | Ribosyldihydronicotinamide dehydrogenase [quinone] |
| 1be1.1.A | 11.11 | monomer | HHblits | NMR | NA | 0.26 | 0.10 | GLUTAMATE MUTASE |
| 1id8.1.A | 11.11 | monomer | HHblits | NMR | NA | 0.26 | 0.10 | METHYLASPARTATE MUTASE S CHAIN |
| 1fmf.1.A | 11.11 | monomer | HHblits | NMR | NA | 0.26 | 0.10 | METHYLASPARTATE MUTASE S CHAIN |
| 1fye.1.A | 8.57 | monomer | HHblits | X-ray | 1.20Å | 0.28 | 0.10 | ASPARTYL DIPEPTIDASE |
| 5k9b.1.A | 14.29 | monomer | HHblits | X-ray | 1.17Å | 0.28 | 0.10 | Flavodoxin-2 |
| 3nu1.1.A | 8.57 | monomer | HHblits | X-ray | 2.50Å | 0.28 | 0.10 | Hemin-binding periplasmic protein |
| 5jb3.1.G | 28.13 | hetero-oligomer | HHblits | EM | NA | 0.35 | 0.09 | 30S ribosomal protein S2 |
| 4v4n.12.A | 28.13 | monomer | HHblits | EM | NA | 0.35 | 0.09 | 30S ribosomal protein S2P |
| 5ig8.1.A | 5.56 | homo-dimer | HHblits | X-ray | 2.28Å | 0.25 | 0.10 | ATP grasp ligase |
| 5ig8.2.A | 5.56 | homo-dimer | HHblits | X-ray | 2.28Å | 0.25 | 0.10 | ATP grasp ligase |
| 2p2s.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.25Å | 0.30 | 0.09 | Putative oxidoreductase |
| 3lyu.1.A | 20.59 | homo-hexamer | HHblits | X-ray | 2.30Å | 0.29 | 0.09 | Putative hydrogenase |
| 1yo6.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.09 | Putative Carbonyl Reductase Sniffer |
| 1yo6.3.B | 17.65 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.09 | Putative Carbonyl Reductase Sniffer |
| 2c1x.1.A | 17.65 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE |
| 4rb4.1.A | 14.71 | homo-12-mer | HHblits | X-ray | 3.88Å | 0.29 | 0.09 | Glycosyltransferase tibC |
| 1xjc.1.A | 13.89 | monomer | HHblits | X-ray | 2.10Å | 0.24 | 0.10 | MobB protein homolog |
| 4i08.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.06Å | 0.31 | 0.09 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG |
| 4eqr.1.A | 18.18 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.09 | Coenzyme A disulfide reductase |
| 1vi5.1.A | 21.88 | monomer | HHblits | X-ray | 2.65Å | 0.33 | 0.09 | 30S ribosomal protein S2P |
| 1vi5.2.A | 21.88 | monomer | HHblits | X-ray | 2.65Å | 0.33 | 0.09 | 30S ribosomal protein S2P |
| 1vi5.4.A | 21.88 | monomer | HHblits | X-ray | 2.65Å | 0.33 | 0.09 | 30S ribosomal protein S2P |
| 3m1l.1.A | 12.12 | homo-dimer | HHblits | X-ray | 2.52Å | 0.30 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3op4.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.30 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 1qfj.1.A | 18.75 | monomer | HHblits | X-ray | 2.20Å | 0.33 | 0.09 | PROTEIN (FLAVIN REDUCTASE) |
| 1qfj.2.A | 18.75 | monomer | HHblits | X-ray | 2.20Å | 0.33 | 0.09 | PROTEIN (FLAVIN REDUCTASE) |
| 1qfj.3.A | 18.75 | monomer | HHblits | X-ray | 2.20Å | 0.33 | 0.09 | PROTEIN (FLAVIN REDUCTASE) |
| 1qfj.4.A | 18.75 | monomer | HHblits | X-ray | 2.20Å | 0.33 | 0.09 | PROTEIN (FLAVIN REDUCTASE) |
| 2acw.1.A | 14.71 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.09 | triterpene UDP-glucosyl transferase UGT71G1 |
| 2acw.1.B | 14.71 | homo-dimer | HHblits | X-ray | 2.60Å | 0.28 | 0.09 | triterpene UDP-glucosyl transferase UGT71G1 |
| 2pq6.1.A | 17.65 | monomer | HHblits | X-ray | 2.10Å | 0.28 | 0.09 | UDP-glucuronosyl/UDP-glucosyltransferase |
| 5cej.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.30 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5cdy.1.C | 21.21 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.30 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 5cdy.1.A | 21.21 | homo-tetramer | HHblits | X-ray | 2.85Å | 0.30 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 3lrx.1.A | 21.21 | homo-hexamer | HHblits | X-ray | 2.60Å | 0.29 | 0.09 | Putative hydrogenase |
| 3kzv.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.09 | Uncharacterized oxidoreductase YIR035C |
| 4v4b.1.C | 28.13 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 5m1j.2.A | 28.13 | monomer | HHblits | EM | 3.30Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 4v7h.1.B | 28.13 | hetero-oligomer | HHblits | EM | 8.90Å | 0.32 | 0.09 | 40S ribosomal protein S0(A) |
| 4v88.1.A | 28.13 | monomer | HHblits | X-ray | 3.00Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 3j77.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0 |
| 4v7r.1.A | 28.13 | monomer | HHblits | X-ray | 4.00Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 4v6i.2.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein rpS0 (S2p) |
| 3j78.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0 |
| 3j6y.43.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0 |
| 3j6x.43.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0 |
| 4v88.76.A | 28.13 | monomer | HHblits | X-ray | 3.00Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 5mc6.16.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 5ll6.1.B | 28.13 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 5jut.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | uS2 (yeast S0) |
| 5juo.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | uS2 (yeast S0) |
| 5juu.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | uS2 (yeast S0) |
| 5jup.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | uS2 (yeast S0) |
| 5jus.46.A | 28.13 | monomer | HHblits | EM | NA | 0.32 | 0.09 | uS2 (yeast S0) |
| 5lyb.1.A | 28.13 | monomer | HHblits | X-ray | 3.25Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 5tga.76.A | 28.13 | monomer | HHblits | X-ray | 3.30Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 4v92.1.C | 28.13 | hetero-oligomer | HHblits | EM | 3.70Å | 0.32 | 0.09 | US2 |
| 4u6f.1.A | 28.13 | monomer | HHblits | X-ray | 3.10Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 4u3n.76.A | 28.13 | monomer | HHblits | X-ray | 3.20Å | 0.32 | 0.09 | 40S ribosomal protein S0-A |
| 4v8m.11.A | 25.00 | monomer | HHblits | EM | 5.57Å | 0.32 | 0.09 | 40S RIBOSOMAL PROTEIN SA, PUTATIVE |
| 3h4i.1.A | 15.15 | monomer | HHblits | X-ray | 1.30Å | 0.29 | 0.09 | Glycosyltransferase GtfA, Glycosyltransferase |
| 5t2a.50.A | 25.00 | monomer | HHblits | EM | NA | 0.32 | 0.09 | uS2 |
| 5myj.1.B | 21.88 | hetero-oligomer | HHblits | EM | 5.60Å | 0.31 | 0.09 | 30S ribosomal protein S2 |
| 4v61.1.B | 31.25 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.09 | Ribosomal Protein S2 |
| 5mmj.1.D | 31.25 | hetero-oligomer | HHblits | EM | 3.65Å | 0.31 | 0.09 | 30S ribosomal protein S2, chloroplastic |
| 5x8r.1.A | 31.25 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.09 | 30S ribosomal protein S2, chloroplastic |
| 3a28.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.09 | L-2.3-butanediol dehydrogenase |
| 1i01.1.D | 18.18 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.29 | 0.09 | BETA-KETOACYL [ACP] REDUCTASE |
| 1q7b.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.29 | 0.09 | 3-oxoacyl-[acyl-carrier protein] reductase |
| 1i01.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.29 | 0.09 | BETA-KETOACYL [ACP] REDUCTASE |
| 5it7.45.A | 25.00 | monomer | HHblits | EM | NA | 0.31 | 0.09 | 40S ribosomal protein S0 |
| 3jaq.1.D | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.09 | uS2 |
| 3j80.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.09 | uS2 |
| 3j81.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.09 | uS2 |
| 4v6w.16.A | 21.88 | monomer | HHblits | EM | NA | 0.31 | 0.09 | 40S ribosomal protein SA |
| 5mrc.41.A | 21.88 | monomer | HHblits | EM | NA | 0.31 | 0.09 | uS2m |
| 5t2u.1.A | 21.88 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 5t2v.1.A | 21.88 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.31 | 0.09 | Oxidoreductase, short chain dehydrogenase/reductase family protein |
| 1ks9.1.A | 25.00 | monomer | HHblits | X-ray | 1.70Å | 0.31 | 0.09 | 2-DEHYDROPANTOATE 2-REDUCTASE |
| 2nm0.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 1.99Å | 0.28 | 0.09 | Probable 3-oxacyl-(Acyl-carrier-protein) reductase |
| 4nkr.1.A | 8.57 | homo-dimer | HHblits | X-ray | 2.41Å | 0.23 | 0.10 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.2.A | 8.57 | homo-dimer | HHblits | X-ray | 2.41Å | 0.23 | 0.10 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.2.B | 8.57 | homo-dimer | HHblits | X-ray | 2.41Å | 0.23 | 0.10 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.3.A | 8.57 | homo-dimer | HHblits | X-ray | 2.41Å | 0.23 | 0.10 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4nkr.3.B | 8.57 | homo-dimer | HHblits | X-ray | 2.41Å | 0.23 | 0.10 | Molybdopterin-guanine dinucleotide biosynthesis protein B |
| 4v9l.2.A | 25.00 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v68.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4w29.2.A | 25.00 | hetero-oligomer | HHblits | X-ray | 3.80Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4w29.1.A | 25.00 | hetero-oligomer | HHblits | X-ray | 3.80Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4k0k.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.40Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9j.2.A | 25.00 | hetero-oligomer | HHblits | X-ray | 3.86Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9k.2.A | 25.00 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 1fjg.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 2om7.1.N | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 3t1h.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.11Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 3oto.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.69Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4dr2.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.25Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4kvb.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 4.20Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4lfa.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.65Å | 0.30 | 0.09 | ribosomal protein S2 |
| 4lf7.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | ribosomal protein S2 |
| 4lfz.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.92Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v8n.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v5q.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v9s.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S Ribosomal Protein S2 |
| 4v5c.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v8c.2.5 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v8h.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S Ribosomal Protein S2 |
| 4v4i.1.8 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.71Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v67.1.E | 25.00 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v4p.1.c | 25.00 | hetero-oligomer | HHblits | X-ray | 5.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v4x.1.D | 25.00 | hetero-oligomer | HHblits | X-ray | 5.00Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9a.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v90.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 2.95Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v9s.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S Ribosomal Protein S2 |
| 4v8f.1.5 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v9b.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v5k.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v5l.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v95.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S Ribosomal Protein S2 |
| 4v63.1.E | 25.00 | hetero-oligomer | HHblits | X-ray | 3.21Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9a.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v9b.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v8u.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.70Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v95.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S Ribosomal Protein S2 |
| 4v4j.1.8 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.83Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v6g.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v5e.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.45Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v8f.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 5j8b.1.8 | 25.00 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4y4p.2.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wzd.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wzo.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wzo.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wr6.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.05Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wr6.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.05Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wsm.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wqr.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el5.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el4.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el6.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el6.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el4.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5e81.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 2.95Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el7.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5e7k.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wu1.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5el5.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.15Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wqu.2.8 | 25.00 | hetero-oligomer | HHblits | X-ray | 2.80Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5imq.1.I | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5imr.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5ibb.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 2.96Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5ibb.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 2.96Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v5a.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 2uua.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 2vqe.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 5lmr.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5lmp.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5lmn.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5lmu.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5lmt.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5lms.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5lmq.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5j4d.1.a | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5w4k.2.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5dox.1.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4z8c.2.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5dox.2.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4zer.1.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4zer.2.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v7p.2.6 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.62Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4gkk.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4gkj.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v4g.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 11.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9q.2.5 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.40Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9i.2.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S Ribosomal protein S2 |
| 4jv5.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.16Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v9h.1.O | 25.00 | hetero-oligomer | HHblits | X-ray | 2.86Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4wt8.2.D | 25.00 | hetero-oligomer | HHblits | X-ray | 3.40Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v4a.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 9.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4xej.2.2 | 25.00 | hetero-oligomer | HHblits | X-ray | 3.80Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5aa0.1.8 | 25.00 | hetero-oligomer | HHblits | EM | 5.00Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 2e5l.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4x62.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.45Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4yhh.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.42Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 5iwa.1.A | 25.00 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 1i94.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.20Å | 0.30 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 2zm6.1.B | 25.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.30 | 0.09 | 30S ribosomal protein S2 |
| 4v5o.1.K | 25.00 | hetero-oligomer | HHblits | X-ray | 3.93Å | 0.30 | 0.09 | RPS0E |
| 3bch.1.A | 21.88 | monomer | HHblits | X-ray | 2.15Å | 0.30 | 0.09 | 40S ribosomal protein SA |
| 3jag.45.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | uS2 |
| 5k0y.1.1 | 21.88 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | ribosomal protein uS2 |
| 4kzx.1.A | 21.88 | hetero-oligomer | HHblits | X-ray | 7.81Å | 0.30 | 0.09 | 40S ribosomal protein SA |
| 4ujc.45.A | 21.88 | monomer | HHblits | EM | 9.50Å | 0.30 | 0.09 | 40S RIBOSOMAL PROTEIN US2 |
| 3j7p.46.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | Ribosomal protein uS2 |
| 4v5z.2.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | 40S Ribosomal protein SA |
| 4ug0.44.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | 40S RIBOSOMAL PROTEIN SA |
| 5aj0.46.A | 21.88 | monomer | HHblits | EM | 3.50Å | 0.30 | 0.09 | 40S ribosomal protein SA |
| 5t2c.66.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | 40S ribosomal protein SA |
| 4ujd.45.A | 21.88 | monomer | HHblits | EM | 8.90Å | 0.30 | 0.09 | 40S RIBOSOMAL PROTEIN US2 |
| 4v6x.16.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | 40S ribosomal protein SA |
| 5a2q.1.C | 21.88 | hetero-oligomer | HHblits | EM | 3.90Å | 0.30 | 0.09 | RIBOSOMAL PROTEIN US2 |
| 5lks.44.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | 40S ribosomal protein SA |
| 5lzz.45.A | 21.88 | monomer | HHblits | EM | 3.47Å | 0.30 | 0.09 | uS2 |
| 3lk7.1.A | 25.00 | monomer | HHblits | X-ray | 1.50Å | 0.30 | 0.09 | UDP-N-acetylmuramoylalanine--D-glutamate ligase |
| 5xxu.1.B | 18.75 | hetero-oligomer | HHblits | EM | 3.35Å | 0.30 | 0.09 | Ribosomal protein uS2 |
| 2acv.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.09 | triterpene UDP-glucosyl transferase UGT71G1 |
| 2acv.1.B | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.09 | triterpene UDP-glucosyl transferase UGT71G1 |
| 3sj7.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 3sj7.1.B | 15.15 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.27 | 0.09 | 3-oxoacyl-(Acyl-carrier-protein) reductase |
| 1uzl.1.A | 15.15 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.09 | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE |
| 2wwv.1.D | 8.82 | hetero-oligomer | HHblits | NMR | NA | 0.25 | 0.09 | N\,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT |
| 5xyi.1.B | 21.88 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.09 | 40S ribosomal protein SA |
| 2uvd.1.A | 18.18 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.09 | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE |
| 5ffq.1.A | 11.76 | homo-dimer | HHblits | X-ray | 2.00Å | 0.25 | 0.09 | ShuY-like protein |
| 4v7e.25.A | 21.88 | monomer | HHblits | EM | NA | 0.30 | 0.09 | 40S ribosomal protein S2 |
| 4v3p.2.A | 21.88 | monomer | HHblits | EM | 34.00Å | 0.30 | 0.09 | 40S ribosomal protein SA |
| 5o5j.1.U | 28.13 | hetero-oligomer | HHblits | EM | 3.45Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3j9w.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein uS2 |
| 5njt.1.B | 25.00 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3jbo.18.A | 21.88 | monomer | HHblits | EM | NA | 0.29 | 0.09 | 40S ribosomal protein uS2 |
| 3jbp.31.A | 21.88 | monomer | HHblits | EM | NA | 0.29 | 0.09 | 40S ribosomal protein uS2 |
| 3jbn.31.A | 21.88 | monomer | HHblits | EM | NA | 0.29 | 0.09 | 40S ribosomal protein uS2 |
| 3j7a.1.C | 21.88 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 40S ribosomal protein uS2 |
| 4v9d.2.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v7s.2.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.25Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9d.1.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v7u.1.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.10Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4u24.1.B | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v69.1.M | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5mgp.1.6 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v5h.1.B | 18.75 | hetero-oligomer | HHblits | EM | 5.80Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 5np6.1.E | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4a2i.1.B | 18.75 | hetero-oligomer | HHblits | EM | 16.50Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 5lze.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5lzb.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5lzc.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 2ykr.1.B | 18.75 | hetero-oligomer | HHblits | EM | 9.80Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 5lzf.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5uyl.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5uyn.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5uyk.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5uyp.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5uyq.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5o2r.1.8 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 6awd.1.G | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 6awc.1.H | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 6awb.1.H | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v4v.1.E | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal subunit protein S2 |
| 5nwy.1.9 | 18.75 | hetero-oligomer | HHblits | EM | 2.93Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4ybb.1.B | 18.75 | hetero-oligomer | HHblits | X-ray | 2.10Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5no3.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v75.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v72.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v70.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v73.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6y.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v71.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6z.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v74.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v77.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v76.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v7a.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v79.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v78.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v5y.1.T | 18.75 | hetero-oligomer | HHblits | X-ray | 4.45Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v4q.1.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.46Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v50.2.C | 18.75 | hetero-oligomer | HHblits | X-ray | 3.22Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v5b.1.7 | 18.75 | hetero-oligomer | HHblits | X-ray | 3.74Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v54.2.S | 18.75 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v4q.2.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.46Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v64.2.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v57.1.R | 18.75 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v5b.2.6 | 18.75 | hetero-oligomer | HHblits | X-ray | 3.74Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v57.2.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v50.1.C | 18.75 | hetero-oligomer | HHblits | X-ray | 3.22Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v56.1.R | 18.75 | hetero-oligomer | HHblits | X-ray | 3.93Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v56.2.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.93Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6q.1.E | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6s.1.b | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6p.1.E | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6r.1.E | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6o.1.E | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6v.1.K | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6m.1.G | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6n.1.c | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v7d.1.9 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3j9z.1.N | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3ja1.1.R | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5h5u.1.c | 18.75 | hetero-oligomer | HHblits | EM | 3.00Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6c.1.A | 18.75 | hetero-oligomer | HHblits | X-ray | 3.19Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v4h.2.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.46Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v4h.1.T | 18.75 | hetero-oligomer | HHblits | X-ray | 3.46Å | 0.29 | 0.09 | 30S RIBOSOMAL PROTEIN S2 |
| 4v6c.2.A | 18.75 | hetero-oligomer | HHblits | X-ray | 3.19Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9o.1.7 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9c.1.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9c.2.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9p.3.7 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9p.2.6 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9o.2.7 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9o.4.6 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9o.3.7 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v9p.4.6 | 18.75 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v66.1.F | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6k.1.c | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v6l.1.F | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4v7i.1.a | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5kcs.1.9 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5kcr.1.7 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5iqr.1.5 | 18.75 | hetero-oligomer | HHblits | EM | 3.00Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3jcn.1.8 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5mdz.1.a | 18.75 | hetero-oligomer | HHblits | EM | 3.10Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3jbv.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3jbu.1.A | 18.75 | hetero-oligomer | HHblits | EM | 3.64Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5u4i.1.6 | 18.75 | hetero-oligomer | HHblits | EM | 3.50Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3jce.1.U | 18.75 | hetero-oligomer | HHblits | EM | 3.20Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3jcd.1.A | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5u9g.1.e | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5u9f.1.e | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5uz4.1.T | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5kps.1.6 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5kpv.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5kpw.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5kpx.1.5 | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4woi.1.B | 18.75 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 4woi.2.6 | 18.75 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 3j9y.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5ju8.1.B | 18.75 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 1uag.1.A | 12.50 | monomer | HHblits | X-ray | 1.95Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYL-L-ALANINE/:D-GLUTAMATE LIGASE |
| 1eeh.1.A | 12.50 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE |
| 2uag.1.A | 12.50 | monomer | HHblits | X-ray | 1.70Å | 0.29 | 0.09 | PROTEIN (UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE) |
| 1e0d.1.A | 12.50 | monomer | HHblits | X-ray | 2.40Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 5a5e.1.A | 12.50 | monomer | HHblits | X-ray | 1.84Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 5a5f.1.A | 12.50 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 1iib.1.A | 12.12 | monomer | HHblits | X-ray | 1.80Å | 0.27 | 0.09 | ENZYME IIB OF THE CELLOBIOSE-SPECIFIC PHOSPHOTRANSFERASE SYSTEM |
| 1e2b.1.A | 12.12 | monomer | HHblits | NMR | NA | 0.27 | 0.09 | ENZYME IIB-CELLOBIOSE |
| 2xpc.1.A | 12.50 | monomer | HHblits | X-ray | 1.49Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 2y67.1.A | 12.50 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.09 | UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE |
| 4jna.1.A | 16.13 | monomer | HHblits | X-ray | 2.00Å | 0.31 | 0.09 | DepH |
| 4jn9.1.A | 16.13 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.09 | DepH |
| 4jn9.1.B | 16.13 | homo-dimer | HHblits | X-ray | 1.90Å | 0.31 | 0.09 | DepH |
| 5li0.1.B | 15.63 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5nd8.1.B | 15.63 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5nd9.1.B | 15.63 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5ngm.1.B | 15.63 | hetero-oligomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 5ng8.1.A | 15.63 | monomer | HHblits | EM | NA | 0.29 | 0.09 | 30S ribosomal protein S2 |
| 2q5c.1.A | 9.68 | monomer | HHblits | X-ray | 1.49Å | 0.31 | 0.09 | NtrC family transcriptional regulator |
| 5jyd.1.A | 18.75 | homo-tetramer | HHblits | X-ray | 1.65Å | 0.28 | 0.09 | Short chain dehydrogenase |
| 1fdr.1.A | 15.63 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.09 | FLAVODOXIN REDUCTASE |
| 1z7e.1.B | 12.12 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.25 | 0.09 | protein ArnA |
| 1z7e.1.A | 12.12 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.25 | 0.09 | protein ArnA |
| 1z7e.1.F | 12.12 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.25 | 0.09 | protein ArnA |
| 4wkg.1.D | 12.12 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.25 | 0.09 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.E | 12.12 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.25 | 0.09 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.F | 12.12 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.25 | 0.09 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.B | 12.12 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.25 | 0.09 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.C | 12.12 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.25 | 0.09 | Bifunctional polymyxin resistance protein ArnA |
| 3fbs.1.A | 16.13 | homo-dimer | HHblits | X-ray | 2.15Å | 0.30 | 0.09 | Oxidoreductase |
| 3ksu.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.09 | 3-oxoacyl-acyl carrier protein reductase |
| 1uls.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.09 | putative 3-oxoacyl-acyl carrier protein reductase |
| 1djq.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.09 | TRIMETHYLAMINE DEHYDROGENASE |
| 1djn.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.09 | TRIMETHYLAMINE DEHYDROGENASE |
| 3c85.1.A | 12.90 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3c85.1.B | 12.90 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3c85.2.A | 12.90 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3c85.2.B | 12.90 | homo-dimer | HHblits | X-ray | 1.90Å | 0.29 | 0.09 | Putative glutathione-regulated potassium-efflux system protein KefB |
| 3guy.1.A | 12.90 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.28 | 0.09 | Short-chain dehydrogenase/reductase SDR |
| 4j34.1.A | 9.68 | monomer | HHblits | X-ray | 2.03Å | 0.28 | 0.09 | Kynurenine 3-monooxygenase |
| 4j34.2.A | 9.68 | monomer | HHblits | X-ray | 2.03Å | 0.28 | 0.09 | Kynurenine 3-monooxygenase |
| 3h7a.1.A | 6.06 | homo-tetramer | HHblits | X-ray | 1.87Å | 0.23 | 0.09 | short chain dehydrogenase |
| 5l76.1.A | 23.33 | monomer | HHblits | X-ray | 2.57Å | 0.30 | 0.08 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 5o1o.1.A | 23.33 | homo-dimer | HHblits | X-ray | 2.48Å | 0.30 | 0.08 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 5l78.1.A | 24.14 | homo-dimer | HHblits | X-ray | 2.68Å | 0.30 | 0.08 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 5l78.1.B | 24.14 | homo-dimer | HHblits | X-ray | 2.68Å | 0.30 | 0.08 | Alpha-aminoadipic semialdehyde synthase, mitochondrial |
| 1pnv.1.A | 20.69 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | GLYCOSYLTRANSFERASE GTFA |
| 1pnv.1.B | 20.69 | homo-dimer | HHblits | X-ray | 2.80Å | 0.30 | 0.08 | GLYCOSYLTRANSFERASE GTFA |
| 3wc4.1.A | 17.24 | monomer | HHblits | X-ray | 1.85Å | 0.29 | 0.08 | UDP-glucose:anthocyanidin 3-O-glucosyltransferase |
| 3ia7.1.A | 13.79 | homo-dimer | HHblits | X-ray | 1.91Å | 0.28 | 0.08 | CalG4 |
| 3ia7.1.B | 13.79 | homo-dimer | HHblits | X-ray | 1.91Å | 0.28 | 0.08 | CalG4 |
| 3otg.1.A | 17.86 | monomer | HHblits | X-ray | 2.08Å | 0.29 | 0.08 | CalG1 |
| 3oth.1.A | 17.86 | monomer | HHblits | X-ray | 2.30Å | 0.29 | 0.08 | CalG1 |
| 5du2.1.A | 23.08 | monomer | HHblits | X-ray | 2.70Å | 0.32 | 0.07 | EspG2 glycosyltransferase |
| 5du2.2.A | 23.08 | monomer | HHblits | X-ray | 2.70Å | 0.32 | 0.07 | EspG2 glycosyltransferase |
| 2c2y.1.A | 11.54 | homo-dimer | HHblits | X-ray | 2.30Å | 0.31 | 0.07 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE-METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE |
| 2c2x.1.B | 11.54 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.07 | METHYLENETETRAHYDROFOLATE DEHYDROGENASE-METHENYLTETRAHYDROFOLATE CYCLOHYDROLASE |
| 1l5x.1.A | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.07 | Survival protein E |
| 1l5x.1.B | 20.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.31 | 0.07 | Survival protein E |
| 2e6h.1.A | 20.00 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6h.1.B | 20.00 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6h.1.C | 20.00 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6h.1.D | 20.00 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 3rsc.1.A | 18.52 | monomer | HHblits | X-ray | 2.19Å | 0.25 | 0.07 | CalG2 |
| 2e6g.1.A | 20.83 | homo-12-mer | HHblits | X-ray | 2.60Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6c.1.B | 20.83 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6c.1.D | 20.83 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e69.1.C | 20.83 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6e.1.A | 20.83 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6e.1.B | 20.83 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6e.1.C | 20.83 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6e.1.D | 20.83 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6c.1.A | 20.83 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2e6g.1.D | 20.83 | homo-12-mer | HHblits | X-ray | 2.60Å | 0.31 | 0.07 | 5'-nucleotidase surE |
| 2ixd.1.A | 25.00 | homo-hexamer | HHblits | X-ray | 1.80Å | 0.36 | 0.06 | LMBE-RELATED PROTEIN |
| 1q74.2.A | 15.00 | monomer | HHblits | X-ray | 1.70Å | 0.32 | 0.06 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) |
| 1q74.1.A | 15.00 | monomer | HHblits | X-ray | 1.70Å | 0.32 | 0.06 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) |
| 4ewl.1.A | 15.00 | homo-dimer | HHblits | X-ray | 1.85Å | 0.32 | 0.06 | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase |
| 4cjx.1.A | 33.33 | homo-dimer | HHblits | X-ray | 2.05Å | 0.40 | 0.05 | C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, PUTATIVE |
| 4cjx.1.B | 33.33 | homo-dimer | HHblits | X-ray | 2.05Å | 0.40 | 0.05 | C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, PUTATIVE |
| 3dfk.1.A | 21.05 | monomer | HHblits | X-ray | 1.80Å | 0.35 | 0.05 | Teicoplanin pseudoaglycone deacetylase Orf2 |
| 1uan.1.A | 15.00 | homo-hexamer | HHblits | X-ray | 2.00Å | 0.27 | 0.06 | hypothetical protein TT1542 |
| 1fqy.1.A | 23.53 | homo-tetramer | HHblits | 2DX | 3.80Å | 0.35 | 0.05 | AQUAPORIN-1 |
| 1ih5.1.A | 23.53 | homo-tetramer | HHblits | 2DX | 3.70Å | 0.35 | 0.05 | AQUAPORIN-1 |
| 1h6i.1.A | 23.53 | monomer | HHblits | 2DX | 3.54Å | 0.35 | 0.05 | AQUAPORIN-1 |
| 1z7e.1.B | 11.11 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.30 | 0.05 | protein ArnA |
| 1z7e.1.A | 11.11 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.30 | 0.05 | protein ArnA |
| 1z7e.1.F | 11.11 | homo-hexamer | HHblits | X-ray | 3.00Å | 0.30 | 0.05 | protein ArnA |
| 4wkg.1.D | 11.11 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.30 | 0.05 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.E | 11.11 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.30 | 0.05 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.F | 11.11 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.30 | 0.05 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.B | 11.11 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.30 | 0.05 | Bifunctional polymyxin resistance protein ArnA |
| 4wkg.1.C | 11.11 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.30 | 0.05 | Bifunctional polymyxin resistance protein ArnA |