 |  | SWISS-MODEL Homology Modelling Report |
Model Building Report
This document lists the results for the homology modelling project "A6VC58" submitted to SWISS-MODEL workspace on Oct. 18, 2017, 4:49 p.m..The submitted primary amino acid sequence is given in Table T1.
If you use any results in your research, please cite the relevant publications:
Marco Biasini; Stefan Bienert; Andrew Waterhouse; Konstantin Arnold; Gabriel Studer; Tobias Schmidt; Florian Kiefer; Tiziano Gallo Cassarino; Martino Bertoni; Lorenza Bordoli; Torsten Schwede. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research (1 July 2014) 42 (W1): W252-W258; doi: 10.1093/nar/gku340.Arnold, K., Bordoli, L., Kopp, J. and Schwede, T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195-201.
Benkert, P., Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343-350
Results
The SWISS-MODEL template library (SMTL version 2017-10-11, PDB release 2017-10-06) was searched with Blast (Altschul et al., 1997) and HHBlits (Remmert, et al., 2011) for evolutionary related structures matching the target sequence in Table T1. For details on the template search, see Materials and Methods. Overall 780 templates were found (Table T2).
Models
The following model was built (see Materials and Methods "Model Building"):
Model #01 | File | Built with | Oligo-State | Ligands | GMQE | QMEAN |
|---|---|---|---|---|---|---|
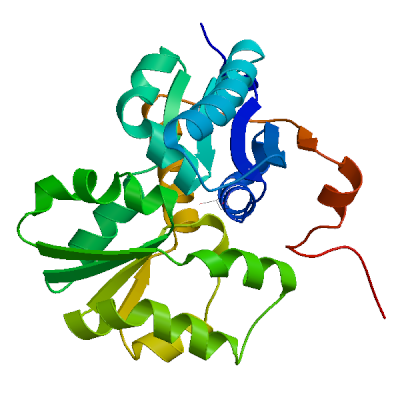 | PDB | ProMod3 Version 1.1.0. | MONOMER (matching prediction) |
1 x DGL: D-GLUTAMIC ACID; | 0.74 | -1.90 |
| 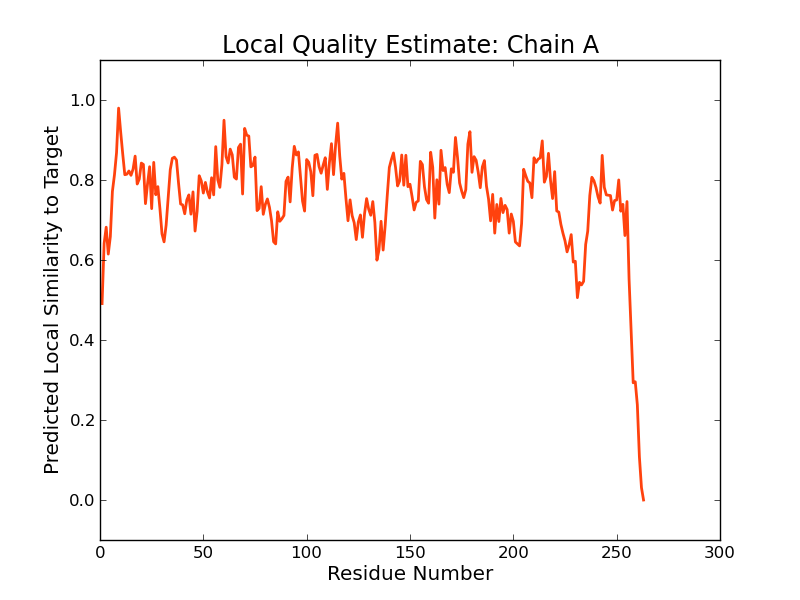 |  |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| 5ijw.1.A | 40.23 | homo-dimer | HHblits | X-ray | 1.76Å | 0.39 | 1 - 263 | 0.98 | Glutamate racemase |
| Ligand | Added to Model | Description | |
|---|---|---|---|
| DGL | ✓ | D-GLUTAMIC ACID | |
| DGL | ✕ - Binding site not conserved. | D-GLUTAMIC ACID | |
| IOD | ✕ - Not biologically relevant. | IODIDE ION | |
| IOD | ✕ - Not biologically relevant. | IODIDE ION | |
| IOD | ✕ - Not biologically relevant. | IODIDE ION |
Target MAAESAPVGVFDSGVGGLSVLREIRARLPAESLLYVADNAHVPYGEKSAEYIRERCERIGDFLLERGAKALVLACNTATA
5ijw.1.A MSDRLAPIGIFDSGVGGLTVARAIIDQLPDEDIVYVGDTGNGPYGPLTIPQIRAHSLAIGDDLVSRGVKALVIACNTASS
Target AAAAELRERYPQ-VPLVAM-EPAVKPAAAATRNGRVGVLATTGTLKSARFAALLDRFASDVQVFTQPCPGLVERIEAGDL
5ijw.1.A ACLRDARERY-SPVPVVEVILPAVRRAVAATRNGRIGVIGTQATIASGAYQDAFAA-ARDTEVFTVACPRFVDFVERGVT
Target HGARTRALLERLLGPILEQGCDTLILGCTHYPFVKPLLAELIPADIAVIDTGAAVARQLERVLSTRALLASG--Q---AA
5ijw.1.A SGRQVLGLAEGYLEPLQLAEVDTLVLGCTHYPMLSGLIQLAMGDNVTLVSSAEETAKDLLRVLTELDLLRPHPDDPSVTA
Target APRFWTSALPEEMERILPILWGSQE-SVGKLDV
5ijw.1.A VRRFEATGDPEAFTALAARFLGPTLDGVRPV--
Materials and Methods
Template Search
Template search with Blast and HHBlits has been performed against the SWISS-MODEL template library (SMTL, last update: 2017-10-11, last included PDB release: 2017-10-06).
The target sequence was searched with BLAST (Altschul et al., 1997) against the primary amino acid sequence contained in the SMTL. A total of 37 templates were found.
An initial HHblits profile has been built using the procedure outlined in (Remmert, et al., 2011), followed by 1 iteration of HHblits against NR20. The obtained profile has then be searched against all profiles of the SMTL. A total of 747 templates were found.
Template Selection
For each identified template, the template's quality has been predicted from features of the target-template alignment. The templates with the highest quality have then been selected for model building.
Model Building
Models are built based on the target-template alignment using ProMod3. Coordinates which are conserved between the target and the template are copied from the template to the model. Insertions and deletions are remodelled using a fragment library. Side chains are then rebuilt. Finally, the geometry of the resulting model is regularized by using a force field. In case loop modelling with ProMod3 fails, an alternative model is built with PROMOD-II (Guex, et al., 1997).
Model Quality Estimation
The global and per-residue model quality has been assessed using the QMEAN scoring function (Benkert, et al., 2011) . For improved performance, weights of the individual QMEAN terms have been trained specifically for SWISS-MODEL.
Ligand Modelling
Ligands present in the template structure are transferred by homology to the model when the following criteria are met (Gallo -Casserino, to be published): (a) The ligands are annotated as biologically relevant in the template library, (b) the ligand is in contact with the model, (c) the ligand is not clashing with the protein, (d) the residues in contact with the ligand are conserved between the target and the template. If any of these four criteria is not satisfied, a certain ligand will not be included in the model. The model summary includes information on why and which ligand has not been included.
Oligomeric State Conservation
Homo-oligomeric structure of the target protein is predicted based on the analysis of pairwise interfaces of the identified template structures. For each relevant interface between polypeptide chains (interfaces with more than 10 residue-residue interactions), the QscoreOligomer (Mariani et al., 2011) is predicted from features such as similarity to target and frequency of observing this interface in the identified templates (Kiefer, Bertoni, Biasini, to be published). The prediction is performed with a random forest regressor using these features as input parameters to predict the probability of conservation for each interface. The QscoreOligomer of the whole complex is then calculated as the weight-averaged QscoreOligomer of the interfaces. The oligomeric state of the target is predicted to be the same as in the template when QscoreOligomer is predicted to be higher or equal to 0.5.
References
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res, 25, 3389-3402.
Remmert, M., Biegert, A., Hauser, A. and Soding, J. (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods, 9, 173-175.
Guex, N. and Peitsch, M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714-2723.
Sali, A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol, 234, 779-815.
Benkert, P., Biasini, M. and Schwede, T. (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343-350.
Mariani, V., Kiefer, F., Schmidt, T., Haas, J. and Schwede, T. (2011) Assessment of template based protein structure predictions in CASP9. Proteins, 79 Suppl 10, 37-58.
Table T1:
Primary amino acid sequence for which templates were searched and models were built.
AVKPAAAATRNGRVGVLATTGTLKSARFAALLDRFASDVQVFTQPCPGLVERIEAGDLHGARTRALLERLLGPILEQGCDTLILGCTHYPFVKPLLAELI
PADIAVIDTGAAVARQLERVLSTRALLASGQAAAPRFWTSALPEEMERILPILWGSQESVGKLDV
Table T2:
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Coverage | Description |
|---|---|---|---|---|---|---|---|---|
| 5ijw.1.A | 40.23 | homo-dimer | HHblits | X-ray | 1.76Å | 0.39 | 0.98 | Glutamate racemase |
| 5ijw.1.B | 40.23 | homo-dimer | HHblits | X-ray | 1.76Å | 0.39 | 0.98 | Glutamate racemase |
| 5hj7.1.A | 38.70 | homo-dimer | HHblits | X-ray | 2.30Å | 0.38 | 0.98 | Glutamate racemase |
| 5hj7.1.A | 41.50 | homo-dimer | BLAST | X-ray | 2.30Å | 0.39 | 0.95 | Glutamate racemase |
| 3hfr.1.A | 34.36 | homo-dimer | HHblits | X-ray | 2.30Å | 0.38 | 0.98 | Glutamate racemase |
| 3hfr.1.B | 34.36 | homo-dimer | HHblits | X-ray | 2.30Å | 0.38 | 0.98 | Glutamate racemase |
| 2jfp.1.A | 32.05 | homo-dimer | HHblits | X-ray | 1.98Å | 0.37 | 0.98 | GLUTAMATE RACEMASE |
| 2jfo.1.B | 32.05 | homo-dimer | HHblits | X-ray | 2.50Å | 0.37 | 0.98 | GLUTAMATE RACEMASE |
| 2jfw.1.A | 33.46 | homo-dimer | HHblits | X-ray | 2.00Å | 0.38 | 0.97 | GLUTAMATE RACEMASE |
| 2vvt.1.A | 32.17 | homo-dimer | HHblits | X-ray | 1.65Å | 0.37 | 0.97 | GLUTAMATE RACEMASE |
| 2vvt.1.B | 32.17 | homo-dimer | HHblits | X-ray | 1.65Å | 0.37 | 0.97 | GLUTAMATE RACEMASE |
| 1zuw.1.A | 32.82 | homo-dimer | HHblits | X-ray | 1.75Å | 0.37 | 0.98 | glutamate racemase 1 |
| 1zuw.1.B | 32.82 | homo-dimer | HHblits | X-ray | 1.75Å | 0.37 | 0.98 | glutamate racemase 1 |
| 2gzm.1.A | 32.05 | homo-dimer | HHblits | X-ray | 1.99Å | 0.37 | 0.98 | Glutamate racemase |
| 2gzm.1.B | 32.05 | homo-dimer | HHblits | X-ray | 1.99Å | 0.37 | 0.98 | Glutamate racemase |
| 2gzm.2.B | 32.05 | homo-dimer | HHblits | X-ray | 1.99Å | 0.37 | 0.98 | Glutamate racemase |
| 2oho.1.A | 31.78 | homo-dimer | HHblits | X-ray | 2.25Å | 0.37 | 0.97 | Glutamate Racemase |
| 2oho.1.B | 31.78 | homo-dimer | HHblits | X-ray | 2.25Å | 0.37 | 0.97 | Glutamate Racemase |
| 2dwu.1.A | 33.72 | homo-dimer | HHblits | X-ray | 1.60Å | 0.37 | 0.97 | Glutamate racemase |
| 2dwu.2.A | 33.72 | homo-dimer | HHblits | X-ray | 1.60Å | 0.37 | 0.97 | Glutamate racemase |
| 2ohv.1.A | 31.91 | homo-dimer | HHblits | X-ray | 2.50Å | 0.37 | 0.97 | Glutamate Racemase |
| 2ohg.1.A | 31.91 | homo-dimer | HHblits | X-ray | 2.50Å | 0.37 | 0.97 | Glutamate racemase |
| 2dwu.1.A | 33.72 | homo-dimer | BLAST | X-ray | 1.60Å | 0.36 | 0.97 | Glutamate racemase |
| 2dwu.2.A | 33.72 | homo-dimer | BLAST | X-ray | 1.60Å | 0.36 | 0.97 | Glutamate racemase |
| 2ohv.1.A | 33.20 | homo-dimer | BLAST | X-ray | 2.50Å | 0.37 | 0.95 | Glutamate Racemase |
| 2ohg.1.A | 33.20 | homo-dimer | BLAST | X-ray | 2.50Å | 0.37 | 0.95 | Glutamate racemase |
| 2oho.1.A | 33.20 | homo-dimer | BLAST | X-ray | 2.25Å | 0.37 | 0.95 | Glutamate Racemase |
| 2oho.1.B | 33.20 | homo-dimer | BLAST | X-ray | 2.25Å | 0.37 | 0.95 | Glutamate Racemase |
| 2jfq.1.A | 32.03 | homo-dimer | HHblits | X-ray | 2.15Å | 0.36 | 0.97 | GLUTAMATE RACEMASE |
| 2jfq.1.B | 32.03 | homo-dimer | HHblits | X-ray | 2.15Å | 0.36 | 0.97 | GLUTAMATE RACEMASE |
| 2jfn.1.A | 40.00 | monomer | HHblits | X-ray | 1.90Å | 0.37 | 0.94 | GLUTAMATE RACEMASE |
| 1zuw.1.A | 34.00 | homo-dimer | BLAST | X-ray | 1.75Å | 0.37 | 0.94 | glutamate racemase 1 |
| 1zuw.1.B | 34.00 | homo-dimer | BLAST | X-ray | 1.75Å | 0.37 | 0.94 | glutamate racemase 1 |
| 2jfn.1.A | 43.21 | monomer | BLAST | X-ray | 1.90Å | 0.39 | 0.92 | GLUTAMATE RACEMASE |
| 2jfw.1.A | 33.60 | homo-dimer | BLAST | X-ray | 2.00Å | 0.38 | 0.93 | GLUTAMATE RACEMASE |
| 3out.1.A | 29.15 | homo-hexamer | HHblits | X-ray | 1.65Å | 0.36 | 0.93 | Glutamate racemase |
| 1b73.1.A | 34.16 | homo-dimer | HHblits | X-ray | 2.30Å | 0.37 | 0.92 | GLUTAMATE RACEMASE |
| 2vvt.1.A | 33.88 | homo-dimer | BLAST | X-ray | 1.65Å | 0.38 | 0.91 | GLUTAMATE RACEMASE |
| 2vvt.1.B | 33.88 | homo-dimer | BLAST | X-ray | 1.65Å | 0.38 | 0.91 | GLUTAMATE RACEMASE |
| 2jfp.1.A | 33.88 | homo-dimer | BLAST | X-ray | 1.98Å | 0.38 | 0.91 | GLUTAMATE RACEMASE |
| 2jfo.1.B | 33.88 | homo-dimer | BLAST | X-ray | 2.50Å | 0.38 | 0.91 | GLUTAMATE RACEMASE |
| 2jfq.1.A | 34.43 | homo-dimer | BLAST | X-ray | 2.15Å | 0.36 | 0.92 | GLUTAMATE RACEMASE |
| 2jfq.1.B | 34.43 | homo-dimer | BLAST | X-ray | 2.15Å | 0.36 | 0.92 | GLUTAMATE RACEMASE |
| 1b73.1.A | 35.29 | homo-dimer | BLAST | X-ray | 2.30Å | 0.38 | 0.90 | GLUTAMATE RACEMASE |
| 3hfr.1.A | 37.34 | homo-dimer | BLAST | X-ray | 2.30Å | 0.39 | 0.88 | Glutamate racemase |
| 3hfr.1.B | 37.34 | homo-dimer | BLAST | X-ray | 2.30Å | 0.39 | 0.88 | Glutamate racemase |
| 3out.1.A | 30.93 | homo-hexamer | BLAST | X-ray | 1.65Å | 0.37 | 0.89 | Glutamate racemase |
| 2gzm.1.A | 32.91 | homo-dimer | BLAST | X-ray | 1.99Å | 0.37 | 0.88 | Glutamate racemase |
| 2gzm.1.B | 32.91 | homo-dimer | BLAST | X-ray | 1.99Å | 0.37 | 0.88 | Glutamate racemase |
| 2gzm.2.B | 32.91 | homo-dimer | BLAST | X-ray | 1.99Å | 0.37 | 0.88 | Glutamate racemase |
| 5ijw.1.A | 44.20 | homo-dimer | BLAST | X-ray | 1.76Å | 0.40 | 0.85 | Glutamate racemase |
| 5ijw.1.B | 44.20 | homo-dimer | BLAST | X-ray | 1.76Å | 0.40 | 0.85 | Glutamate racemase |
| 3uho.1.A | 31.51 | monomer | BLAST | X-ray | 2.20Å | 0.35 | 0.90 | Glutamate racemase |
| 3uhf.2.A | 31.51 | monomer | BLAST | X-ray | 1.83Å | 0.35 | 0.90 | Glutamate racemase |
| 3uhp.1.A | 31.51 | monomer | BLAST | X-ray | 2.79Å | 0.35 | 0.90 | Glutamate racemase |
| 2jfy.1.B | 26.12 | homo-dimer | HHblits | X-ray | 1.90Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 2jfz.1.A | 26.12 | homo-dimer | HHblits | X-ray | 1.86Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 2jfx.1.A | 26.12 | homo-dimer | HHblits | X-ray | 2.30Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 2jfx.1.B | 26.12 | homo-dimer | HHblits | X-ray | 2.30Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 2jfy.1.A | 26.12 | homo-dimer | HHblits | X-ray | 1.90Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 4b1f.1.B | 26.12 | homo-dimer | HHblits | X-ray | 2.05Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 2w4i.1.A | 26.12 | homo-tetramer | HHblits | X-ray | 1.87Å | 0.33 | 0.92 | GLUTAMATE RACEMASE |
| 3uho.1.A | 28.51 | monomer | HHblits | X-ray | 2.20Å | 0.34 | 0.91 | Glutamate racemase |
| 3uhf.2.A | 28.51 | monomer | HHblits | X-ray | 1.83Å | 0.34 | 0.91 | Glutamate racemase |
| 3uhp.1.A | 28.51 | monomer | HHblits | X-ray | 2.79Å | 0.34 | 0.91 | Glutamate racemase |
| 5w1q.1.A | 26.12 | homo-dimer | HHblits | X-ray | 1.95Å | 0.33 | 0.92 | Glutamate racemase |
| 5b19.1.A | 23.08 | homo-dimer | HHblits | X-ray | 1.85Å | 0.31 | 0.78 | Aspartate racemase |
| 5b19.1.B | 23.08 | homo-dimer | HHblits | X-ray | 1.85Å | 0.31 | 0.78 | Aspartate racemase |
| 1jfl.1.A | 24.51 | homo-dimer | HHblits | X-ray | 1.90Å | 0.32 | 0.77 | ASPARTATE RACEMASE |
| 2dx7.1.A | 24.02 | homo-dimer | HHblits | X-ray | 2.00Å | 0.32 | 0.77 | aspartate racemase |
| 2dx7.1.B | 24.02 | homo-dimer | HHblits | X-ray | 2.00Å | 0.32 | 0.77 | aspartate racemase |
| 3s81.1.A | 20.59 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.77 | Putative aspartate racemase |
| 5ell.1.A | 18.84 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.78 | Asp/Glu_racemase family protein |
| 5ell.1.B | 18.84 | homo-dimer | HHblits | X-ray | 1.80Å | 0.30 | 0.78 | Asp/Glu_racemase family protein |
| 2eq5.1.A | 21.26 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.78 | 228aa long hypothetical hydantoin racemase |
| 2eq5.2.B | 21.26 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.78 | 228aa long hypothetical hydantoin racemase |
| 5hqt.1.A | 18.45 | homo-dimer | HHblits | X-ray | 1.60Å | 0.29 | 0.78 | aspartate/glutamate racemase |
| 4fq7.1.A | 15.31 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.28 | 0.79 | Maleate cis-trans isomerase |
| 3ojc.1.A | 19.21 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.30 | 0.77 | Putative aspartate/glutamate racemase |
| 3ojc.1.B | 19.21 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.30 | 0.77 | Putative aspartate/glutamate racemase |
| 3ojc.1.C | 19.21 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.30 | 0.77 | Putative aspartate/glutamate racemase |
| 3ojc.1.D | 19.21 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.30 | 0.77 | Putative aspartate/glutamate racemase |
| 5elm.1.A | 17.56 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.77 | Asp/Glu_racemase family protein |
| 5elm.1.B | 17.56 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.77 | Asp/Glu_racemase family protein |
| 5elm.2.A | 17.56 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.77 | Asp/Glu_racemase family protein |
| 5elm.2.B | 17.56 | homo-dimer | HHblits | X-ray | 2.00Å | 0.29 | 0.77 | Asp/Glu_racemase family protein |
| 4fq5.1.A | 14.35 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.79 | Maleate cis-trans isomerase |
| 4fq5.1.B | 14.35 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.79 | Maleate cis-trans isomerase |
| 2zsk.1.A | 19.31 | homo-dimer | HHblits | X-ray | 2.55Å | 0.30 | 0.76 | 226aa long hypothetical aspartate racemase |
| 2dgd.1.A | 14.42 | homo-octamer | HHblits | X-ray | 2.90Å | 0.27 | 0.78 | 223aa long hypothetical arylmalonate decarboxylase |
| 2xed.1.A | 14.49 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.78 | PUTATIVE MALEATE ISOMERASE |
| 2xec.1.A | 15.05 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.78 | PUTATIVE MALEATE ISOMERASE |
| 3qvj.1.A | 18.32 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.28 | 0.76 | Putative hydantoin racemase |
| 4ix1.1.A | 13.94 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.78 | hypothetical protein |
| 4ix1.1.B | 13.94 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.78 | hypothetical protein |
| 2jfy.1.B | 33.70 | homo-dimer | BLAST | X-ray | 1.90Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 2jfz.1.A | 33.70 | homo-dimer | BLAST | X-ray | 1.86Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 2jfx.1.A | 33.70 | homo-dimer | BLAST | X-ray | 2.30Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 2jfx.1.B | 33.70 | homo-dimer | BLAST | X-ray | 2.30Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 2jfy.1.A | 33.70 | homo-dimer | BLAST | X-ray | 1.90Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 4b1f.1.B | 33.70 | homo-dimer | BLAST | X-ray | 2.05Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 2w4i.1.A | 33.70 | homo-tetramer | BLAST | X-ray | 1.87Å | 0.37 | 0.68 | GLUTAMATE RACEMASE |
| 3qvl.1.A | 17.33 | homo-hexamer | HHblits | X-ray | 1.82Å | 0.28 | 0.76 | Putative hydantoin racemase |
| 4rs3.1.A | 15.92 | monomer | HHblits | X-ray | 1.40Å | 0.28 | 0.76 | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein |
| 5w1q.1.A | 31.15 | homo-dimer | BLAST | X-ray | 1.95Å | 0.35 | 0.69 | Glutamate racemase |
| 4yv7.1.A | 13.86 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.76 | Periplasmic binding protein/LacI transcriptional regulator |
| 5lfd.1.A | 15.50 | homo-hexamer | HHblits | X-ray | 2.15Å | 0.27 | 0.75 | Allantoin racemase |
| 4wzz.1.A | 9.36 | monomer | HHblits | X-ray | 1.70Å | 0.26 | 0.77 | Putative sugar ABC transporter, substrate-binding protein |
| 3ixm.1.A | 17.09 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.75 | Arylmalonate decarboxylase |
| 3hzr.1.A | 12.31 | homo-dimer | HHblits | X-ray | 3.00Å | 0.28 | 0.74 | Tryptophanyl-tRNA synthetase |
| 5ix8.1.A | 11.33 | monomer | HHblits | X-ray | 1.60Å | 0.25 | 0.77 | Putative sugar ABC transport system, substrate-binding protein |
| 3dg9.1.A | 16.58 | monomer | HHblits | X-ray | 1.50Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3dtv.3.A | 16.58 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3dtv.2.A | 16.58 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3dtv.1.A | 16.58 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3dtv.4.A | 16.58 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3eis.1.A | 16.58 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3eis.2.A | 16.58 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 2vlb.1.A | 16.58 | monomer | HHblits | X-ray | 1.92Å | 0.26 | 0.75 | ARYLMALONATE DECARBOXYLASE |
| 2vlb.2.A | 16.58 | monomer | HHblits | X-ray | 1.92Å | 0.26 | 0.75 | ARYLMALONATE DECARBOXYLASE |
| 2vlb.3.A | 16.58 | monomer | HHblits | X-ray | 1.92Å | 0.26 | 0.75 | ARYLMALONATE DECARBOXYLASE |
| 2vlb.4.A | 16.58 | monomer | HHblits | X-ray | 1.92Å | 0.26 | 0.75 | ARYLMALONATE DECARBOXYLASE |
| 3k9c.1.A | 13.64 | homo-dimer | HHblits | X-ray | 2.14Å | 0.26 | 0.75 | Transcriptional regulator, LacI family protein |
| 3ixl.1.A | 16.16 | monomer | HHblits | X-ray | 1.45Å | 0.26 | 0.75 | Arylmalonate decarboxylase |
| 3ctp.1.A | 11.73 | homo-dimer | HHblits | X-ray | 1.41Å | 0.26 | 0.74 | Periplasmic binding protein/LacI transcriptional regulator |
| 2q5c.1.A | 10.12 | monomer | HHblits | X-ray | 1.49Å | 0.26 | 0.63 | NtrC family transcriptional regulator |
| 4ccz.1.A | 18.12 | monomer | HHblits | X-ray | 2.70Å | 0.29 | 0.52 | METHIONINE SYNTHASE |
| 5dmm.1.A | 18.98 | monomer | HHblits | X-ray | 1.78Å | 0.27 | 0.52 | Homocysteine S-methyltransferase |
| 5dmn.1.A | 18.98 | homo-dimer | HHblits | X-ray | 2.89Å | 0.27 | 0.52 | Homocysteine S-methyltransferase |
| 4f06.1.A | 15.67 | monomer | HHblits | X-ray | 1.30Å | 0.28 | 0.51 | Extracellular ligand-binding receptor |
| 3obb.1.A | 20.31 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.29 | 0.48 | Probable 3-hydroxyisobutyrate dehydrogenase |
| 5a9r.1.A | 18.75 | homo-dimer | HHblits | X-ray | 1.55Å | 0.28 | 0.48 | IMINE REDUCTASE |
| 5a9s.1.A | 18.75 | hetero-oligomer | HHblits | X-ray | 2.06Å | 0.28 | 0.48 | IMINE REDUCTASE |
| 5fwn.1.B | 18.75 | homo-dimer | HHblits | X-ray | 2.14Å | 0.28 | 0.48 | IMINE REDUCTASE |
| 3pef.1.A | 13.28 | homo-tetramer | HHblits | X-ray | 2.07Å | 0.27 | 0.48 | 6-phosphogluconate dehydrogenase, NAD-binding |
| 3pef.2.C | 13.28 | homo-tetramer | HHblits | X-ray | 2.07Å | 0.27 | 0.48 | 6-phosphogluconate dehydrogenase, NAD-binding |
| 4mwa.1.A | 16.80 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.47 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.1.B | 16.80 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.47 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.1.C | 16.80 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.47 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.1.D | 16.80 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.47 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.2.B | 16.80 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.47 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.2.D | 16.80 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.28 | 0.47 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4fx7.1.A | 14.29 | monomer | HHblits | X-ray | 2.08Å | 0.27 | 0.48 | Imidazole glycerol phosphate synthase subunit HisF |
| 3gbv.1.A | 10.40 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.47 | Putative LacI-family transcriptional regulator |
| 3gbv.1.B | 10.40 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.47 | Putative LacI-family transcriptional regulator |
| 3tdn.1.A | 14.40 | homo-dimer | HHblits | X-ray | 1.40Å | 0.26 | 0.47 | FLR SYMMETRIC ALPHA-BETA TIM BARREL |
| 4wxe.1.A | 17.36 | monomer | HHblits | X-ray | 1.50Å | 0.29 | 0.46 | Transcriptional regulator, LacI family |
| 4axk.1.A | 17.36 | monomer | HHblits | X-ray | 2.25Å | 0.27 | 0.46 | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE |
| 4axk.2.A | 17.36 | monomer | HHblits | X-ray | 2.25Å | 0.27 | 0.46 | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE |
| 4rk3.1.A | 18.49 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.45 | Transcriptional regulator, LacI family |
| 4ywh.1.A | 9.68 | monomer | HHblits | X-ray | 2.35Å | 0.25 | 0.47 | ABC TRANSPORTER SOLUTE BINDING PROTEIN |
| 4wd0.1.A | 16.67 | homo-hexamer | HHblits | X-ray | 1.50Å | 0.27 | 0.45 | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase |
| 5dn1.1.A | 19.33 | monomer | HHblits | X-ray | 1.95Å | 0.28 | 0.45 | Phosphoribosyl isomerase A |
| 4rk5.1.A | 17.65 | homo-dimer | HHblits | X-ray | 1.35Å | 0.28 | 0.45 | Transcriptional regulator, LacI family |
| 2y88.1.A | 16.39 | monomer | HHblits | X-ray | 1.33Å | 0.26 | 0.46 | PHOSPHORIBOSYL ISOMERASE A |
| 2y89.1.A | 16.39 | monomer | HHblits | X-ray | 2.50Å | 0.26 | 0.46 | PHOSPHORIBOSYL ISOMERASE A |
| 2y85.1.A | 16.39 | monomer | HHblits | X-ray | 2.40Å | 0.26 | 0.46 | PHOSPHORIBOSYL ISOMERASE A |
| 3zs4.1.A | 16.39 | monomer | HHblits | X-ray | 1.90Å | 0.26 | 0.46 | PHOSPHORIBOSYL ISOMERASE A |
| 2x30.1.A | 18.49 | monomer | HHblits | X-ray | 1.95Å | 0.27 | 0.45 | PHOSPHORIBOSYL ISOMERASE A |
| 3lft.1.A | 14.41 | monomer | HHblits | X-ray | 1.35Å | 0.28 | 0.45 | uncharacterized protein |
| 4u28.1.A | 16.81 | monomer | HHblits | X-ray | 1.33Å | 0.27 | 0.45 | Phosphoribosyl isomerase A |
| 2qu7.1.A | 12.71 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.45 | Putative transcriptional regulator |
| 5ide.1.B | 9.92 | hetero-oligomer | HHblits | EM | NA | 0.25 | 0.46 | Glutamate receptor 3 |
| 5ide.1.D | 9.92 | hetero-oligomer | HHblits | EM | NA | 0.25 | 0.46 | Glutamate receptor 3 |
| 5idf.1.B | 9.92 | hetero-oligomer | HHblits | EM | NA | 0.25 | 0.46 | Glutamate receptor 3 |
| 5idf.1.D | 9.92 | hetero-oligomer | HHblits | EM | NA | 0.25 | 0.46 | Glutamate receptor 3 |
| 3bbl.1.A | 13.45 | homo-dimer | HHblits | X-ray | 2.35Å | 0.27 | 0.45 | Regulatory protein of LacI family |
| 3e61.1.A | 10.17 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.45 | Putative transcriptional repressor of ribose operon |
| 3g85.1.A | 11.02 | monomer | HHblits | X-ray | 1.84Å | 0.26 | 0.45 | Transcriptional regulator (LacI family) |
| 2lbp.1.A | 13.91 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.43 | LEUCINE-BINDING PROTEIN |
| 4evs.1.A | 14.78 | monomer | HHblits | X-ray | 1.45Å | 0.28 | 0.43 | PUTATIVE ABC TRANSPORTER SUBUNIT, SUBSTRATE-BINDING COMPONENT |
| 3g1u.1.A | 14.29 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.25 | 0.45 | Adenosylhomocysteinase |
| 1z16.1.A | 13.04 | monomer | HHblits | X-ray | 1.72Å | 0.28 | 0.43 | Leu/Ile/Val-binding protein |
| 1z15.1.A | 13.04 | monomer | HHblits | X-ray | 1.70Å | 0.28 | 0.43 | Leu/Ile/Val-binding protein |
| 1iu9.1.A | 24.27 | monomer | HHblits | X-ray | 2.04Å | 0.32 | 0.39 | aspartate racemase |
| 2iyo.1.A | 12.04 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.41 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2eq5.1.A | 15.38 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.39 | 228aa long hypothetical hydantoin racemase |
| 2eq5.2.B | 15.38 | homo-dimer | HHblits | X-ray | 2.20Å | 0.29 | 0.39 | 228aa long hypothetical hydantoin racemase |
| 2w8z.1.A | 11.21 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.40 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 1h7x.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.01Å | 0.28 | 0.40 | DIHYDROPYRIMIDINE DEHYDROGENASE |
| 1jfl.1.A | 21.78 | homo-dimer | HHblits | X-ray | 1.90Å | 0.30 | 0.38 | ASPARTATE RACEMASE |
| 2iz1.1.B | 12.15 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.40 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2iz1.1.A | 12.15 | homo-dimer | HHblits | X-ray | 2.30Å | 0.26 | 0.40 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 3s81.1.A | 14.56 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.39 | Putative aspartate racemase |
| 2dx7.1.A | 20.79 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.38 | aspartate racemase |
| 2dx7.1.B | 20.79 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.38 | aspartate racemase |
| 2jfq.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.15Å | 0.29 | 0.38 | GLUTAMATE RACEMASE |
| 2jfq.1.B | 17.65 | homo-dimer | HHblits | X-ray | 2.15Å | 0.29 | 0.38 | GLUTAMATE RACEMASE |
| 2pn1.1.A | 12.26 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.40 | Carbamoylphosphate synthase large subunit |
| 3ojc.1.A | 9.62 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.27 | 0.39 | Putative aspartate/glutamate racemase |
| 3ojc.1.B | 9.62 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.27 | 0.39 | Putative aspartate/glutamate racemase |
| 3ojc.1.C | 9.62 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.27 | 0.39 | Putative aspartate/glutamate racemase |
| 3ojc.1.D | 9.62 | homo-tetramer | HHblits | X-ray | 1.75Å | 0.27 | 0.39 | Putative aspartate/glutamate racemase |
| 5hj7.1.A | 18.81 | homo-dimer | HHblits | X-ray | 2.30Å | 0.29 | 0.38 | Glutamate racemase |
| 5hqt.1.A | 12.62 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.39 | aspartate/glutamate racemase |
| 5elm.1.A | 12.62 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.39 | Asp/Glu_racemase family protein |
| 5elm.1.B | 12.62 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.39 | Asp/Glu_racemase family protein |
| 5elm.2.A | 12.62 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.39 | Asp/Glu_racemase family protein |
| 5elm.2.B | 12.62 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.39 | Asp/Glu_racemase family protein |
| 3qvl.1.A | 18.45 | homo-hexamer | HHblits | X-ray | 1.82Å | 0.27 | 0.39 | Putative hydantoin racemase |
| 5ell.1.A | 11.65 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.39 | Asp/Glu_racemase family protein |
| 5ell.1.B | 11.65 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.39 | Asp/Glu_racemase family protein |
| 5ijw.1.A | 18.81 | homo-dimer | HHblits | X-ray | 1.76Å | 0.28 | 0.38 | Glutamate racemase |
| 5ijw.1.B | 18.81 | homo-dimer | HHblits | X-ray | 1.76Å | 0.28 | 0.38 | Glutamate racemase |
| 2gzm.1.A | 15.84 | homo-dimer | HHblits | X-ray | 1.99Å | 0.28 | 0.38 | Glutamate racemase |
| 2gzm.1.B | 15.84 | homo-dimer | HHblits | X-ray | 1.99Å | 0.28 | 0.38 | Glutamate racemase |
| 2gzm.2.B | 15.84 | homo-dimer | HHblits | X-ray | 1.99Å | 0.28 | 0.38 | Glutamate racemase |
| 3hfr.1.A | 14.85 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.38 | Glutamate racemase |
| 3hfr.1.B | 14.85 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.38 | Glutamate racemase |
| 3g0o.1.A | 15.84 | homo-dimer | HHblits | X-ray | 1.80Å | 0.28 | 0.38 | 3-hydroxyisobutyrate dehydrogenase |
| 3pdu.1.A | 17.00 | homo-tetramer | HHblits | X-ray | 1.89Å | 0.28 | 0.38 | 3-hydroxyisobutyrate dehydrogenase family protein |
| 3q3c.1.A | 17.00 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.28 | 0.38 | Probable 3-hydroxyisobutyrate dehydrogenase |
| 2zsk.1.A | 7.84 | homo-dimer | HHblits | X-ray | 2.55Å | 0.26 | 0.38 | 226aa long hypothetical aspartate racemase |
| 2oho.1.A | 14.85 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.38 | Glutamate Racemase |
| 2oho.1.B | 14.85 | homo-dimer | HHblits | X-ray | 2.25Å | 0.27 | 0.38 | Glutamate Racemase |
| 5lfd.1.A | 19.39 | homo-hexamer | HHblits | X-ray | 2.15Å | 0.29 | 0.37 | Allantoin racemase |
| 3out.1.A | 15.00 | homo-hexamer | HHblits | X-ray | 1.65Å | 0.28 | 0.38 | Glutamate racemase |
| 1zuw.1.A | 14.00 | homo-dimer | HHblits | X-ray | 1.75Å | 0.28 | 0.38 | glutamate racemase 1 |
| 1zuw.1.B | 14.00 | homo-dimer | HHblits | X-ray | 1.75Å | 0.28 | 0.38 | glutamate racemase 1 |
| 2dwu.1.A | 14.85 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.38 | Glutamate racemase |
| 2dwu.2.A | 14.85 | homo-dimer | HHblits | X-ray | 1.60Å | 0.27 | 0.38 | Glutamate racemase |
| 3lp8.1.A | 16.16 | monomer | HHblits | X-ray | 2.15Å | 0.28 | 0.37 | Phosphoribosylamine-glycine ligase |
| 2jfw.1.A | 14.00 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.38 | GLUTAMATE RACEMASE |
| 2vvt.1.A | 11.88 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.38 | GLUTAMATE RACEMASE |
| 2vvt.1.B | 11.88 | homo-dimer | HHblits | X-ray | 1.65Å | 0.27 | 0.38 | GLUTAMATE RACEMASE |
| 2jfp.1.A | 10.89 | homo-dimer | HHblits | X-ray | 1.98Å | 0.26 | 0.38 | GLUTAMATE RACEMASE |
| 2jfo.1.B | 10.89 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.38 | GLUTAMATE RACEMASE |
| 2ohv.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.38 | Glutamate Racemase |
| 2ohg.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.50Å | 0.27 | 0.38 | Glutamate racemase |
| 5b19.1.A | 11.88 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.38 | Aspartate racemase |
| 5b19.1.B | 11.88 | homo-dimer | HHblits | X-ray | 1.85Å | 0.26 | 0.38 | Aspartate racemase |
| 5w1q.1.A | 11.00 | homo-dimer | HHblits | X-ray | 1.95Å | 0.26 | 0.38 | Glutamate racemase |
| 3uho.1.A | 15.00 | monomer | HHblits | X-ray | 2.20Å | 0.25 | 0.38 | Glutamate racemase |
| 3uhf.2.A | 15.00 | monomer | HHblits | X-ray | 1.83Å | 0.25 | 0.38 | Glutamate racemase |
| 3uhp.1.A | 15.00 | monomer | HHblits | X-ray | 2.79Å | 0.25 | 0.38 | Glutamate racemase |
| 2vlb.1.A | 15.31 | monomer | HHblits | X-ray | 1.92Å | 0.27 | 0.37 | ARYLMALONATE DECARBOXYLASE |
| 2vlb.2.A | 15.31 | monomer | HHblits | X-ray | 1.92Å | 0.27 | 0.37 | ARYLMALONATE DECARBOXYLASE |
| 2vlb.3.A | 15.31 | monomer | HHblits | X-ray | 1.92Å | 0.27 | 0.37 | ARYLMALONATE DECARBOXYLASE |
| 2vlb.4.A | 15.31 | monomer | HHblits | X-ray | 1.92Å | 0.27 | 0.37 | ARYLMALONATE DECARBOXYLASE |
| 3dg9.1.A | 15.31 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.37 | Arylmalonate decarboxylase |
| 2gpw.1.A | 6.80 | monomer | HHblits | X-ray | 2.20Å | 0.23 | 0.39 | Biotin carboxylase |
| 2gpw.4.A | 6.80 | monomer | HHblits | X-ray | 2.20Å | 0.23 | 0.39 | Biotin carboxylase |
| 2dgd.1.A | 14.43 | homo-octamer | HHblits | X-ray | 2.90Å | 0.26 | 0.37 | 223aa long hypothetical arylmalonate decarboxylase |
| 2jfn.1.A | 15.79 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.36 | GLUTAMATE RACEMASE |
| 2w90.1.A | 11.46 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.36 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2w90.1.B | 11.46 | homo-dimer | HHblits | X-ray | 2.20Å | 0.26 | 0.36 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2xec.1.A | 10.31 | homo-dimer | HHblits | X-ray | 2.20Å | 0.25 | 0.37 | PUTATIVE MALEATE ISOMERASE |
| 3lzc.1.A | 17.39 | homo-dimer | HHblits | X-ray | 2.26Å | 0.29 | 0.35 | Dph2 |
| 3lzc.1.B | 17.39 | homo-dimer | HHblits | X-ray | 2.26Å | 0.29 | 0.35 | Dph2 |
| 3ixm.1.A | 13.54 | monomer | HHblits | X-ray | 1.90Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 3dtv.3.A | 13.68 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 3dtv.2.A | 13.68 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 3dtv.1.A | 13.68 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 3dtv.4.A | 13.68 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 3eis.1.A | 13.68 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 3eis.2.A | 13.68 | monomer | HHblits | X-ray | 2.10Å | 0.26 | 0.36 | Arylmalonate decarboxylase |
| 2xed.1.A | 8.25 | monomer | HHblits | X-ray | 1.95Å | 0.24 | 0.37 | PUTATIVE MALEATE ISOMERASE |
| 5mlk.1.A | 18.48 | homo-dimer | HHblits | X-ray | 1.94Å | 0.28 | 0.35 | Acetyl-COA carboxylase |
| 3ixl.1.A | 11.46 | monomer | HHblits | X-ray | 1.45Å | 0.24 | 0.36 | Arylmalonate decarboxylase |
| 2bdq.1.A | 19.78 | homo-dimer | HHblits | X-ray | 2.30Å | 0.28 | 0.34 | copper homeostasis protein CutC |
| 2yl2.1.A | 16.48 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.34 | ACETYL-COA CARBOXYLASE 1 |
| 2yl2.2.A | 16.48 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.34 | ACETYL-COA CARBOXYLASE 1 |
| 3u9s.1.A | 22.22 | hetero-oligomer | HHblits | X-ray | 3.50Å | 0.29 | 0.34 | Methylcrotonyl-CoA carboxylase, alpha-subunit |
| 3u9t.1.A | 22.22 | hetero-oligomer | HHblits | X-ray | 2.90Å | 0.29 | 0.34 | Methylcrotonyl-CoA carboxylase, alpha-subunit |
| 3d4o.1.A | 18.48 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.35 | Dipicolinate synthase subunit A |
| 3d4o.1.B | 18.48 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.35 | Dipicolinate synthase subunit A |
| 1ulz.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.34 | pyruvate carboxylase n-terminal domain |
| 5dou.1.A | 16.85 | homo-dimer | HHblits | X-ray | 2.60Å | 0.29 | 0.34 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
| 5dot.1.A | 16.85 | homo-dimer | HHblits | X-ray | 2.40Å | 0.29 | 0.34 | Carbamoyl-phosphate synthase [ammonia], mitochondrial |
| 3gid.1.A | 15.38 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.34 | Acetyl-CoA carboxylase 2 |
| 3gid.2.A | 15.38 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.34 | Acetyl-CoA carboxylase 2 |
| 3glk.1.A | 15.38 | monomer | HHblits | X-ray | 2.10Å | 0.27 | 0.34 | Acetyl-CoA carboxylase 2 |
| 5kkn.2.A | 15.38 | monomer | HHblits | X-ray | 2.60Å | 0.27 | 0.34 | Acetyl-CoA carboxylase 2 |
| 5kkn.1.A | 15.38 | monomer | HHblits | X-ray | 2.60Å | 0.27 | 0.34 | Acetyl-CoA carboxylase 2 |
| 2gps.1.A | 17.39 | monomer | HHblits | X-ray | 2.80Å | 0.26 | 0.35 | Biotin carboxylase |
| 1iu9.1.A | 19.77 | monomer | HHblits | X-ray | 2.04Å | 0.32 | 0.32 | aspartate racemase |
| 2c00.1.A | 17.39 | monomer | HHblits | X-ray | 2.50Å | 0.26 | 0.35 | BIOTIN CARBOXYLASE |
| 2c00.2.A | 17.39 | monomer | HHblits | X-ray | 2.50Å | 0.26 | 0.35 | BIOTIN CARBOXYLASE |
| 2vqd.1.A | 17.39 | monomer | HHblits | X-ray | 2.41Å | 0.26 | 0.35 | BIOTIN CARBOXYLASE |
| 1nvm.1.B | 13.19 | hetero-oligomer | HHblits | X-ray | 1.70Å | 0.27 | 0.34 | acetaldehyde dehydrogenase (acylating) |
| 1nvm.2.B | 13.19 | hetero-oligomer | HHblits | X-ray | 1.70Å | 0.27 | 0.34 | acetaldehyde dehydrogenase (acylating) |
| 2gpw.1.A | 17.58 | monomer | HHblits | X-ray | 2.20Å | 0.27 | 0.34 | Biotin carboxylase |
| 2gpw.4.A | 17.58 | monomer | HHblits | X-ray | 2.20Å | 0.27 | 0.34 | Biotin carboxylase |
| 4mv4.2.A | 18.68 | monomer | HHblits | X-ray | 1.61Å | 0.27 | 0.34 | Biotin carboxylase |
| 1ez1.1.A | 10.53 | homo-dimer | HHblits | X-ray | 1.75Å | 0.23 | 0.36 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE 2 |
| 1ez1.1.B | 10.53 | homo-dimer | HHblits | X-ray | 1.75Å | 0.23 | 0.36 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE 2 |
| 1gso.1.A | 9.78 | monomer | HHblits | X-ray | 1.60Å | 0.26 | 0.35 | PROTEIN (GLYCINAMIDE RIBONUCLEOTIDE SYNTHETASE) |
| 3rv4.1.A | 17.78 | homo-dimer | HHblits | X-ray | 1.98Å | 0.27 | 0.34 | Biotin carboxylase |
| 2vpq.1.A | 13.48 | homo-dimer | HHblits | X-ray | 2.10Å | 0.28 | 0.34 | ACETYL-COA CARBOXYLASE |
| 3mjf.1.A | 8.79 | monomer | HHblits | X-ray | 1.47Å | 0.26 | 0.34 | Phosphoribosylamine--glycine ligase |
| 2qk4.1.A | 13.19 | monomer | HHblits | X-ray | 2.45Å | 0.26 | 0.34 | Trifunctional purine biosynthetic protein adenosine-3 |
| 3ouu.1.A | 12.22 | homo-dimer | HHblits | X-ray | 2.25Å | 0.26 | 0.34 | Biotin carboxylase |
| 3g8d.1.A | 17.98 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.34 | Biotin carboxylase |
| 3ktd.1.A | 15.38 | homo-dimer | HHblits | X-ray | 2.60Å | 0.25 | 0.34 | Prephenate dehydrogenase |
| 1mio.1.B | 10.00 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.26 | 0.34 | NITROGENASE MOLYBDENUM IRON PROTEIN (BETA CHAIN) |
| 1mio.1.D | 10.00 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.26 | 0.34 | NITROGENASE MOLYBDENUM IRON PROTEIN (BETA CHAIN) |
| 4wn9.1.B | 10.00 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.26 | 0.34 | Nitrogenase molybdenum-iron protein beta chain |
| 3wgi.1.B | 14.61 | homo-dimer | HHblits | X-ray | 3.25Å | 0.27 | 0.34 | Redox-sensing transcriptional repressor rex |
| 3wg9.2.A | 14.61 | homo-dimer | HHblits | X-ray | 1.97Å | 0.27 | 0.34 | Redox-sensing transcriptional repressor rex |
| 3wg9.2.B | 14.61 | homo-dimer | HHblits | X-ray | 1.97Å | 0.27 | 0.34 | Redox-sensing transcriptional repressor rex |
| 3wgh.1.B | 14.61 | homo-dimer | HHblits | X-ray | 2.05Å | 0.27 | 0.34 | Redox-sensing transcriptional repressor rex |
| 3wgg.1.A | 14.61 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.34 | Redox-sensing transcriptional repressor rex |
| 3wgg.1.B | 14.61 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.34 | Redox-sensing transcriptional repressor rex |
| 3lp8.1.A | 15.73 | monomer | HHblits | X-ray | 2.15Å | 0.27 | 0.34 | Phosphoribosylamine-glycine ligase |
| 1dv2.1.A | 16.67 | monomer | HHblits | X-ray | 2.50Å | 0.26 | 0.34 | BIOTIN CARBOXYLASE |
| 2ip4.1.A | 16.85 | monomer | HHblits | X-ray | 2.80Å | 0.27 | 0.34 | Phosphoribosylamine--glycine ligase |
| 2ip4.2.A | 16.85 | monomer | HHblits | X-ray | 2.80Å | 0.27 | 0.34 | Phosphoribosylamine--glycine ligase |
| 3g8c.1.A | 17.05 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.33 | Biotin carboxylase |
| 3g8c.2.A | 17.05 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.33 | Biotin carboxylase |
| 2w6m.1.A | 17.05 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.33 | BIOTIN CARBOXYLASE |
| 1bnc.1.B | 17.05 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.33 | BIOTIN CARBOXYLASE |
| 1bnc.1.A | 17.05 | homo-dimer | HHblits | X-ray | 2.40Å | 0.27 | 0.33 | BIOTIN CARBOXYLASE |
| 4hr7.1.A | 17.05 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.27 | 0.33 | Biotin carboxylase |
| 4hr7.1.C | 17.05 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.27 | 0.33 | Biotin carboxylase |
| 4hr7.1.E | 17.05 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.27 | 0.33 | Biotin carboxylase |
| 4hr7.1.F | 17.05 | hetero-oligomer | HHblits | X-ray | 2.50Å | 0.27 | 0.33 | Biotin carboxylase |
| 2j9g.2.A | 17.05 | monomer | HHblits | X-ray | 2.05Å | 0.27 | 0.33 | BIOTIN CARBOXYLASE |
| 2vr1.2.A | 17.05 | monomer | HHblits | X-ray | 2.60Å | 0.27 | 0.33 | BIOTIN CARBOXYLASE |
| 5h80.1.A | 12.22 | homo-dimer | HHblits | X-ray | 1.70Å | 0.25 | 0.34 | Carboxylase |
| 3rv3.1.A | 14.77 | homo-dimer | HHblits | X-ray | 1.91Å | 0.27 | 0.33 | Biotin carboxylase |
| 5e38.1.B | 25.61 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.32 | 0.31 | Uracil phosphoribosyltransferase |
| 5e38.1.A | 25.61 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.32 | 0.31 | Uracil phosphoribosyltransferase |
| 5e38.1.D | 25.61 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.32 | 0.31 | Uracil phosphoribosyltransferase |
| 2pn1.1.A | 11.36 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.33 | Carbamoylphosphate synthase large subunit |
| 2yya.1.A | 13.79 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.33 | Phosphoribosylamine--glycine ligase |
| 2ys6.1.A | 12.36 | monomer | HHblits | X-ray | 2.21Å | 0.26 | 0.34 | Phosphoribosylglycinamide synthetase |
| 5uhw.1.A | 11.63 | homo-dimer | HHblits | X-ray | 2.24Å | 0.28 | 0.32 | Oxidoreductase protein |
| 2ehj.1.A | 23.17 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.32 | 0.31 | Uracil phosphoribosyltransferase |
| 2ehj.1.C | 23.17 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.32 | 0.31 | Uracil phosphoribosyltransferase |
| 2hjs.1.A | 16.28 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.32 | USG-1 protein homolog |
| 2xcl.1.A | 7.95 | monomer | HHblits | X-ray | 2.10Å | 0.25 | 0.33 | PHOSPHORIBOSYLAMINE--GLYCINE LIGASE |
| 3trh.1.A | 13.95 | homo-octamer | HHblits | X-ray | 2.20Å | 0.27 | 0.32 | Phosphoribosylaminoimidazole carboxylase carboxyltransferase subunit |
| 1i5e.1.A | 20.73 | homo-dimer | HHblits | X-ray | 3.00Å | 0.31 | 0.31 | URACIL PHOSPHORIBOSYLTRANSFERASE |
| 1ez1.1.A | 13.95 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.32 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE 2 |
| 1ez1.1.B | 13.95 | homo-dimer | HHblits | X-ray | 1.75Å | 0.27 | 0.32 | PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE 2 |
| 4f3y.1.A | 17.86 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.32 | Dihydrodipicolinate reductase |
| 1o5o.1.A | 21.69 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.29 | 0.31 | Uracil phosphoribosyltransferase |
| 5tov.1.A | 13.79 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.26 | 0.33 | Adenosylhomocysteinase |
| 5tov.1.B | 13.79 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.26 | 0.33 | Adenosylhomocysteinase |
| 4grd.1.A | 20.48 | homo-octamer | HHblits | X-ray | 1.85Å | 0.29 | 0.31 | Phosphoribosylaminoimidazole carboxylase catalytic subunit |
| 3opq.1.A | 19.28 | homo-octamer | HHblits | X-ray | 2.00Å | 0.29 | 0.31 | Phosphoribosylaminoimidazole carboxylase,catalytic subunit |
| 3oow.1.A | 19.28 | homo-octamer | HHblits | X-ray | 1.75Å | 0.29 | 0.31 | Phosphoribosylaminoimidazole carboxylase,catalytic subunit |
| 3lp6.1.B | 16.67 | homo-octamer | HHblits | X-ray | 1.70Å | 0.28 | 0.32 | Phosphoribosylaminoimidazole carboxylase catalytic subunit |
| 3lp6.1.A | 16.67 | homo-octamer | HHblits | X-ray | 1.70Å | 0.28 | 0.32 | Phosphoribosylaminoimidazole carboxylase catalytic subunit |
| 1v9s.1.B | 20.48 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.31 | uracil phosphoribosyltransferase |
| 1v9s.1.A | 20.48 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.31 | uracil phosphoribosyltransferase |
| 1v9s.1.D | 20.48 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.29 | 0.31 | uracil phosphoribosyltransferase |
| 1xmp.1.A | 16.87 | homo-octamer | HHblits | X-ray | 1.80Å | 0.29 | 0.31 | phosphoribosylaminoimidazole carboxylase |
| 3btv.1.A | 22.22 | homo-dimer | HHblits | X-ray | 2.10Å | 0.31 | 0.31 | Galactose/lactose metabolism regulatory protein GAL80 |
| 3bts.1.A | 22.22 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.31 | 0.31 | Galactose/lactose metabolism regulatory protein GAL80 |
| 3bts.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.31 | 0.31 | Galactose/lactose metabolism regulatory protein GAL80 |
| 2ipl.1.A | 24.39 | monomer | HHblits | X-ray | 1.20Å | 0.30 | 0.31 | D-galactose-binding periplasmic protein |
| 5ere.1.A | 21.95 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.31 | Extracellular ligand-binding receptor |
| 3ors.1.A | 15.66 | homo-octamer | HHblits | X-ray | 1.45Å | 0.28 | 0.31 | N5-Carboxyaminoimidazole Ribonucleotide Mutase |
| 3ors.1.B | 15.66 | homo-octamer | HHblits | X-ray | 1.45Å | 0.28 | 0.31 | N5-Carboxyaminoimidazole Ribonucleotide Mutase |
| 3ors.1.E | 15.66 | homo-octamer | HHblits | X-ray | 1.45Å | 0.28 | 0.31 | N5-Carboxyaminoimidazole Ribonucleotide Mutase |
| 3gdo.1.A | 9.41 | homo-dimer | HHblits | X-ray | 2.03Å | 0.26 | 0.32 | Uncharacterized oxidoreductase yvaA |
| 3s99.1.A | 14.29 | monomer | HHblits | X-ray | 2.05Å | 0.27 | 0.32 | Basic membrane lipoprotein |
| 3rcb.1.A | 19.51 | monomer | HHblits | X-ray | 2.49Å | 0.29 | 0.31 | Sugar 3-ketoreductase |
| 4iil.1.A | 11.90 | monomer | HHblits | X-ray | 1.30Å | 0.27 | 0.32 | Membrane lipoprotein TpN38(b) |
| 4wd3.1.A | 6.90 | homo-dimer | HHblits | X-ray | 2.80Å | 0.24 | 0.33 | L-amino acid ligase |
| 5kws.1.A | 23.46 | monomer | HHblits | X-ray | 1.32Å | 0.30 | 0.31 | Galactose-binding protein |
| 1glg.1.A | 24.69 | monomer | HHblits | X-ray | 2.00Å | 0.30 | 0.31 | GALACTOSE/GLUCOSE-BINDING PROTEIN |
| 2qw1.1.A | 24.69 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.31 | D-galactose-binding periplasmic protein |
| 2fvy.1.A | 24.69 | monomer | HHblits | X-ray | 0.92Å | 0.30 | 0.31 | D-galactose-binding periplasmic protein |
| 2ipm.1.A | 24.69 | monomer | HHblits | X-ray | 1.12Å | 0.30 | 0.31 | D-galactose-binding periplasmic protein |
| 2ipn.1.A | 24.69 | monomer | HHblits | X-ray | 1.15Å | 0.29 | 0.31 | D-galactose-binding periplasmic protein |
| 2hph.1.A | 23.46 | monomer | HHblits | X-ray | 1.33Å | 0.29 | 0.31 | D-galactose-binding periplasmic protein |
| 2ys6.1.A | 13.10 | monomer | HHblits | X-ray | 2.21Å | 0.26 | 0.32 | Phosphoribosylglycinamide synthetase |
| 1h6c.1.A | 10.71 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.26 | 0.32 | PRECURSOR FORM OF GLUCOSE-FRUCTOSE OXIDOREDUCTASE |
| 2jfy.1.B | 10.59 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 2jfz.1.A | 10.59 | homo-dimer | HHblits | X-ray | 1.86Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 2jfx.1.A | 10.59 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 2jfx.1.B | 10.59 | homo-dimer | HHblits | X-ray | 2.30Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 2jfy.1.A | 10.59 | homo-dimer | HHblits | X-ray | 1.90Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 4b1f.1.B | 10.59 | homo-dimer | HHblits | X-ray | 2.05Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 2w4i.1.A | 10.59 | homo-tetramer | HHblits | X-ray | 1.87Å | 0.25 | 0.32 | GLUTAMATE RACEMASE |
| 4iil.1.A | 19.75 | monomer | HHblits | X-ray | 1.30Å | 0.29 | 0.31 | Membrane lipoprotein TpN38(b) |
| 3cea.1.A | 10.84 | homo-tetramer | HHblits | X-ray | 2.40Å | 0.27 | 0.31 | Myo-inositol 2-dehydrogenase |
| 4n54.1.A | 13.58 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.28 | 0.31 | Inositol dehydrogenase |
| 4n54.1.C | 13.58 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.28 | 0.31 | Inositol dehydrogenase |
| 4n54.1.D | 13.58 | homo-tetramer | HHblits | X-ray | 2.05Å | 0.28 | 0.31 | Inositol dehydrogenase |
| 1xtu.1.A | 8.54 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.27 | 0.31 | Probable uracil phosphoribosyltransferase |
| 3g6w.1.A | 8.54 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.27 | 0.31 | Uracil phosphoribosyltransferase |
| 3g6w.1.B | 8.54 | homo-tetramer | HHblits | X-ray | 2.90Å | 0.27 | 0.31 | Uracil phosphoribosyltransferase |
| 1xtv.1.B | 8.54 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.27 | 0.31 | Probable uracil phosphoribosyltransferase |
| 1xtv.1.C | 8.54 | homo-tetramer | HHblits | X-ray | 2.60Å | 0.27 | 0.31 | Probable uracil phosphoribosyltransferase |
| 1xtt.1.A | 8.54 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.27 | 0.31 | Probable uracil phosphoribosyltransferase |
| 1xtt.1.B | 8.54 | homo-tetramer | HHblits | X-ray | 1.80Å | 0.27 | 0.31 | Probable uracil phosphoribosyltransferase |
| 1vst.1.A | 8.54 | homo-tetramer | HHblits | X-ray | 2.80Å | 0.27 | 0.31 | Uracil phosphoribosyltransferase |
| 3rc9.1.A | 17.28 | monomer | HHblits | X-ray | 1.91Å | 0.28 | 0.31 | Sugar 3-ketoreductase |
| 2e55.1.A | 13.41 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.27 | 0.31 | Uracil phosphoribosyltransferase |
| 4yle.1.A | 14.63 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.31 | Periplasmic binding protein/LacI transcriptional regulator |
| 2xcl.1.A | 11.90 | monomer | HHblits | X-ray | 2.10Å | 0.25 | 0.32 | PHOSPHORIBOSYLAMINE--GLYCINE LIGASE |
| 3rc1.1.A | 17.50 | monomer | HHblits | X-ray | 1.71Å | 0.28 | 0.30 | Sugar 3-ketoreductase |
| 3q2k.1.A | 16.46 | homo-octamer | HHblits | X-ray | 2.13Å | 0.29 | 0.30 | oxidoreductase |
| 3q2k.1.B | 16.46 | homo-octamer | HHblits | X-ray | 2.13Å | 0.29 | 0.30 | oxidoreductase |
| 5ktk.1.A | 13.58 | monomer | HHblits | X-ray | 1.98Å | 0.27 | 0.31 | Polyketide synthase PksJ |
| 4iq0.1.A | 9.88 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.31 | Oxidoreductase, Gfo/Idh/MocA family |
| 4iq0.2.B | 9.88 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.31 | Oxidoreductase, Gfo/Idh/MocA family |
| 2hqb.1.A | 16.25 | monomer | HHblits | X-ray | 2.70Å | 0.28 | 0.30 | Transcriptional activator of comK gene |
| 3gbp.1.A | 9.76 | monomer | HHblits | X-ray | 2.40Å | 0.26 | 0.31 | GALACTOSE-BINDING PROTEIN |
| 4z0n.1.A | 13.75 | monomer | HHblits | X-ray | 1.26Å | 0.28 | 0.30 | Periplasmic binding protein/LacI transcriptional regulator |
| 5a02.1.A | 8.64 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.31 | ALDOSE-ALDOSE OXIDOREDUCTASE |
| 3mjf.1.A | 16.05 | monomer | HHblits | X-ray | 1.47Å | 0.26 | 0.31 | Phosphoribosylamine--glycine ligase |
| 2qk4.1.A | 11.11 | monomer | HHblits | X-ray | 2.45Å | 0.26 | 0.31 | Trifunctional purine biosynthetic protein adenosine-3 |
| 4ru1.1.A | 11.39 | monomer | HHblits | X-ray | 1.50Å | 0.27 | 0.30 | Monosaccharide ABC transporter substrate-binding protein, CUT2 family |
| 3q2i.1.A | 19.74 | homo-octamer | HHblits | X-ray | 1.50Å | 0.30 | 0.29 | dehydrogenase |
| 3q2i.1.A | 12.82 | homo-octamer | HHblits | X-ray | 1.50Å | 0.27 | 0.29 | dehydrogenase |
| 3rc7.1.A | 19.48 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.29 | Sugar 3-ketoreductase |
| 4d5g.1.A | 13.75 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.25 | 0.30 | CYCLOHEXANE-1,2-DIONE HYDROLASE |
| 3x43.1.A | 18.42 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.29 | 0.29 | O-ureido-L-serine synthase |
| 3x43.1.B | 18.42 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.29 | 0.29 | O-ureido-L-serine synthase |
| 4iq0.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.29 | Oxidoreductase, Gfo/Idh/MocA family |
| 4iq0.2.B | 14.29 | homo-dimer | HHblits | X-ray | 2.00Å | 0.27 | 0.29 | Oxidoreductase, Gfo/Idh/MocA family |
| 3x44.1.A | 14.10 | homo-tetramer | HHblits | X-ray | 1.90Å | 0.26 | 0.29 | O-ureido-L-serine synthase |
| 2rdm.1.A | 15.58 | homo-hexamer | HHblits | X-ray | 1.76Å | 0.27 | 0.29 | Response regulator receiver protein |
| 2rdm.1.B | 15.58 | homo-hexamer | HHblits | X-ray | 1.76Å | 0.27 | 0.29 | Response regulator receiver protein |
| 2rdm.1.C | 15.58 | homo-hexamer | HHblits | X-ray | 1.76Å | 0.27 | 0.29 | Response regulator receiver protein |
| 3rcb.1.A | 17.11 | monomer | HHblits | X-ray | 2.49Å | 0.28 | 0.29 | Sugar 3-ketoreductase |
| 2pgn.1.A | 13.92 | homo-tetramer | HHblits | X-ray | 1.20Å | 0.25 | 0.30 | Cyclohexane-1,2-dione Hydrolase (Cdh) |
| 5eqv.1.A | 14.10 | monomer | HHblits | X-ray | 1.45Å | 0.26 | 0.29 | bifunctional 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase periplasmic precursor protein |
| 5vev.1.A | 10.26 | monomer | HHblits | X-ray | 1.90Å | 0.25 | 0.29 | Phosphoribosylamine--glycine ligase |
| 2lle.1.A | 11.84 | monomer | HHblits | NMR | NA | 0.27 | 0.29 | Chemotaxis protein CheY, Imidazole glycerol phosphate synthase subunit HisF chimera |
| 5uhw.1.A | 11.69 | homo-dimer | HHblits | X-ray | 2.24Å | 0.26 | 0.29 | Oxidoreductase protein |
| 4mkx.1.A | 15.07 | homo-tetramer | HHblits | X-ray | 1.60Å | 0.30 | 0.28 | Inositol dehydrogenase |
| 3cwo.1.A | 9.21 | homo-dimer | HHblits | X-ray | 3.10Å | 0.26 | 0.29 | beta/alpha-barrel protein based on 1THF and 1TMY |
| 1gso.1.A | 16.00 | monomer | HHblits | X-ray | 1.60Å | 0.27 | 0.28 | PROTEIN (GLYCINAMIDE RIBONUCLEOTIDE SYNTHETASE) |
| 4mge.1.A | 13.70 | monomer | HHblits | X-ray | 1.85Å | 0.28 | 0.28 | PTS system, cellobiose-specific IIB component |
| 4mge.2.A | 13.70 | monomer | HHblits | X-ray | 1.85Å | 0.28 | 0.28 | PTS system, cellobiose-specific IIB component |
| 3gka.1.A | 13.16 | monomer | HHblits | X-ray | 2.30Å | 0.24 | 0.29 | N-ethylmaleimide reductase |
| 4egj.1.A | 20.55 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.28 | D-alanine--D-alanine ligase |
| 1qh1.1.B | 15.49 | hetero-oligomer | HHblits | X-ray | 1.60Å | 0.29 | 0.27 | PROTEIN (NITROGENASE MOLYBDENUM IRON PROTEIN) |
| 3jyf.1.A | 13.70 | monomer | HHblits | X-ray | 2.43Å | 0.26 | 0.28 | 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein |
| 3ec7.1.A | 14.71 | homo-tetramer | HHblits | X-ray | 2.15Å | 0.31 | 0.26 | Putative Dehydrogenase |
| 3lft.1.A | 12.68 | monomer | HHblits | X-ray | 1.35Å | 0.28 | 0.27 | uncharacterized protein |
| 3nkl.1.A | 15.71 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.26 | UDP-D-quinovosamine 4-dehydrogenase |
| 3nkl.1.B | 15.71 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.26 | UDP-D-quinovosamine 4-dehydrogenase |
| 4by9.1.D | 12.50 | hetero-oligomer | HHblits | NMR | NA | 0.26 | 0.27 | 50S RIBOSOMAL PROTEIN L7AE |
| 4by9.1.G | 12.50 | hetero-oligomer | HHblits | NMR | NA | 0.26 | 0.27 | 50S RIBOSOMAL PROTEIN L7AE |
| 4by9.1.J | 12.50 | hetero-oligomer | HHblits | NMR | NA | 0.26 | 0.27 | 50S RIBOSOMAL PROTEIN L7AE |
| 4by9.1.M | 12.50 | hetero-oligomer | HHblits | NMR | NA | 0.26 | 0.27 | 50S RIBOSOMAL PROTEIN L7AE |
| 3t9p.1.A | 14.08 | homo-dimer | HHblits | X-ray | 1.97Å | 0.27 | 0.27 | Mandelate racemase/muconate lactonizing enzyme family protein |
| 3j9t.1.C | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9t.1.E | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9t.1.G | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9u.1.C | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9u.1.E | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9u.1.G | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9v.1.E | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9v.1.G | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 3j9v.1.I | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 4iyj.1.A | 15.94 | homo-dimer | HHblits | X-ray | 1.37Å | 0.29 | 0.26 | GDSL-like protein |
| 5bw9.2.G | 20.90 | hetero-oligomer | HHblits | X-ray | 7.00Å | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 5voz.1.A | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voz.1.E | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voy.1.A | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voy.1.C | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voz.1.C | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5d80.1.C | 20.90 | hetero-oligomer | HHblits | X-ray | 6.20Å | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 5vox.1.A | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5vox.1.C | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5bw9.2.H | 20.90 | hetero-oligomer | HHblits | X-ray | 7.00Å | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A |
| 5voy.1.E | 20.90 | hetero-oligomer | HHblits | EM | NA | 0.31 | 0.25 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 3qg1.1.A | 21.21 | monomer | HHblits | X-ray | 2.95Å | 0.32 | 0.25 | V-type ATP synthase alpha chain |
| 2qui.1.B | 7.04 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 2quk.1.A | 7.04 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 2qui.1.A | 7.04 | homo-dimer | HHblits | X-ray | 2.40Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 5cx1.1.B | 11.43 | hetero-oligomer | HHblits | X-ray | 1.75Å | 0.27 | 0.26 | Nitrogenase molybdenum-iron protein beta chain |
| 1g21.1.B | 11.43 | hetero-oligomer | HHblits | X-ray | 3.00Å | 0.27 | 0.26 | NITROGENASE MOLYBDENUM-IRON PROTEIN BETA CHAIN |
| 1m34.1.B | 11.43 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.27 | 0.26 | Nitrogenase Molybdenum-Iron Protein beta chain |
| 1ulh.1.A | 7.04 | homo-dimer | HHblits | X-ray | 2.31Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 1ulh.1.B | 7.04 | homo-dimer | HHblits | X-ray | 2.31Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 1r6u.1.A | 7.04 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 1r6u.1.B | 7.04 | homo-dimer | HHblits | X-ray | 2.00Å | 0.26 | 0.27 | Tryptophanyl-tRNA synthetase |
| 4xpi.1.B | 11.59 | hetero-oligomer | HHblits | X-ray | 1.97Å | 0.27 | 0.26 | Nitrogenase molybdenum-iron protein beta chain |
| 4hf7.1.A | 13.43 | homo-dimer | HHblits | X-ray | 1.77Å | 0.29 | 0.25 | Putative acylhydrolase |
| 1yzf.1.A | 16.42 | monomer | HHblits | X-ray | 1.90Å | 0.28 | 0.25 | lipase/acylhydrolase |
| 3q2k.1.A | 23.08 | homo-octamer | HHblits | X-ray | 2.13Å | 0.31 | 0.25 | oxidoreductase |
| 3q2k.1.B | 23.08 | homo-octamer | HHblits | X-ray | 2.13Å | 0.31 | 0.25 | oxidoreductase |
| 3e9m.1.A | 7.25 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.26 | Oxidoreductase, Gfo/Idh/MocA family |
| 3e9m.1.B | 7.25 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.26 | Oxidoreductase, Gfo/Idh/MocA family |
| 3e9m.2.A | 7.25 | homo-dimer | HHblits | X-ray | 2.70Å | 0.26 | 0.26 | Oxidoreductase, Gfo/Idh/MocA family |
| 3btv.1.A | 13.24 | homo-dimer | HHblits | X-ray | 2.10Å | 0.27 | 0.26 | Galactose/lactose metabolism regulatory protein GAL80 |
| 3bts.1.A | 13.24 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.27 | 0.26 | Galactose/lactose metabolism regulatory protein GAL80 |
| 3bts.1.B | 13.24 | hetero-oligomer | HHblits | X-ray | 2.70Å | 0.27 | 0.26 | Galactose/lactose metabolism regulatory protein GAL80 |
| 1j00.1.A | 13.43 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.25 | Thioesterase I |
| 1u8u.1.A | 13.43 | monomer | HHblits | X-ray | 2.08Å | 0.28 | 0.25 | Acyl-CoA thioesterase I |
| 1ryd.1.A | 10.29 | homo-tetramer | HHblits | X-ray | 2.20Å | 0.26 | 0.26 | glucose-fructose oxidoreductase |
| 1rye.1.B | 10.29 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.26 | glucose-fructose oxidoreductase |
| 1rye.1.A | 10.29 | homo-tetramer | HHblits | X-ray | 2.30Å | 0.26 | 0.26 | glucose-fructose oxidoreductase |
| 4jj6.1.A | 10.14 | homo-octamer | HHblits | X-ray | 1.80Å | 0.25 | 0.26 | Acetyl xylan esterase |
| 4ppy.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.25 | Putative acylhydrolase |
| 1ofg.1.A | 10.45 | homo-tetramer | HHblits | X-ray | 2.70Å | 0.27 | 0.25 | GLUCOSE-FRUCTOSE OXIDOREDUCTASE |
| 1evj.1.A | 11.94 | homo-dimer | HHblits | X-ray | 2.70Å | 0.27 | 0.25 | GLUCOSE-FRUCTOSE OXIDOREDUCTASE |
| 5b5s.1.A | 13.24 | monomer | HHblits | X-ray | 1.50Å | 0.25 | 0.26 | Acetic acid |
| 3hp4.1.A | 8.96 | monomer | HHblits | X-ray | 1.35Å | 0.26 | 0.25 | GDSL-esterase |
| 1v2g.1.A | 13.64 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.25 | Acyl-CoA thioesterase I |
| 4jj4.1.A | 10.29 | homo-octamer | HHblits | X-ray | 2.13Å | 0.25 | 0.26 | Acetyl xylan esterase |
| 3rc7.1.A | 16.92 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.25 | Sugar 3-ketoreductase |
| 2w90.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2w90.1.B | 14.29 | homo-dimer | HHblits | X-ray | 2.20Å | 0.30 | 0.24 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2ho5.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.56Å | 0.28 | 0.24 | Oxidoreductase, Gfo/Idh/MocA family |
| 3i05.1.A | 9.52 | homo-dimer | HHblits | X-ray | 2.80Å | 0.27 | 0.24 | Tryptophanyl-tRNA synthetase |
| 1o5t.1.A | 7.94 | homo-dimer | HHblits | X-ray | 2.50Å | 0.26 | 0.24 | Tryptophanyl-tRNA synthetase |
| 2ake.1.B | 7.94 | homo-dimer | HHblits | X-ray | 3.10Å | 0.26 | 0.24 | Tryptophanyl-tRNA synthetase |
| 3tze.1.A | 9.52 | homo-dimer | HHblits | X-ray | 2.60Å | 0.26 | 0.24 | Tryptophanyl-tRNA synthetase |
| 1b73.1.A | 12.90 | homo-dimer | HHblits | X-ray | 2.30Å | 0.27 | 0.23 | GLUTAMATE RACEMASE |
| 3pbk.1.A | 11.29 | monomer | HHblits | X-ray | 3.00Å | 0.27 | 0.23 | Fatty Acyl-Adenylate Ligase |
| 1r6t.1.A | 7.94 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.24 | Tryptophanyl-tRNA synthetase |
| 1r6t.1.B | 7.94 | homo-dimer | HHblits | X-ray | 2.10Å | 0.26 | 0.24 | Tryptophanyl-tRNA synthetase |
| 2azx.1.B | 7.94 | homo-dimer | HHblits | X-ray | 2.80Å | 0.26 | 0.24 | Tryptophanyl-tRNA synthetase |
| 4lda.1.A | 19.67 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.23 | TadZ |
| 4lda.3.A | 19.67 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.23 | TadZ |
| 4lda.4.A | 19.67 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.23 | TadZ |
| 4lda.5.A | 19.67 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.23 | TadZ |
| 2p2q.1.A | 12.90 | monomer | HHblits | X-ray | 2.42Å | 0.26 | 0.23 | Acetyl-coenzyme A synthetase |
| 2p2f.1.A | 11.29 | monomer | HHblits | X-ray | 2.58Å | 0.25 | 0.23 | Acetyl-coenzyme A synthetase |
| 2p2m.1.A | 11.29 | monomer | HHblits | X-ray | 2.11Å | 0.25 | 0.23 | Acetyl-coenzyme A synthetase |
| 1pg3.1.A | 11.29 | monomer | HHblits | X-ray | 2.30Å | 0.25 | 0.23 | acetyl-CoA synthetase |
| 4mg4.1.A | 11.48 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.23 | Phosphonomutase |
| 4mg4.2.B | 11.48 | homo-dimer | HHblits | X-ray | 1.70Å | 0.24 | 0.23 | Phosphonomutase |
| 1wau.1.A | 17.54 | homo-trimer | HHblits | X-ray | 2.80Å | 0.27 | 0.22 | KHG/KDPG ALDOLASE |
| 4nae.1.B | 26.42 | hetero-oligomer | HHblits | X-ray | 2.00Å | 0.33 | 0.20 | Heptaprenylglyceryl phosphate synthase |
| 3noy.1.A | 25.93 | homo-dimer | HHblits | X-ray | 2.70Å | 0.31 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 3noy.2.A | 25.93 | homo-dimer | HHblits | X-ray | 2.70Å | 0.31 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 5bw9.2.G | 18.52 | hetero-oligomer | HHblits | X-ray | 7.00Å | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 5voz.1.A | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voz.1.E | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voy.1.A | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voy.1.C | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5voz.1.C | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5d80.1.C | 18.52 | hetero-oligomer | HHblits | X-ray | 6.20Å | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 5vox.1.A | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5vox.1.C | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 5bw9.2.H | 18.52 | hetero-oligomer | HHblits | X-ray | 7.00Å | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 5voy.1.E | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A |
| 3j9t.1.C | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9t.1.E | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9t.1.G | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9u.1.C | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9u.1.E | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9u.1.G | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9v.1.E | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9v.1.G | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3j9v.1.I | 18.52 | hetero-oligomer | HHblits | EM | NA | 0.30 | 0.20 | V-type proton ATPase catalytic subunit A |
| 3w3a.1.A | 20.75 | hetero-oligomer | HHblits | X-ray | 3.90Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 3w3a.1.B | 20.75 | hetero-oligomer | HHblits | X-ray | 3.90Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 3w3a.1.C | 20.75 | hetero-oligomer | HHblits | X-ray | 3.90Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 5gar.1.A | 20.75 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 5gar.1.B | 20.75 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 5gar.1.C | 20.75 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 5gas.1.A | 20.75 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 5gas.1.C | 20.75 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 5gas.1.B | 20.75 | hetero-oligomer | HHblits | EM | NA | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 3a5d.1.A | 20.75 | hetero-oligomer | HHblits | X-ray | 4.80Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 3a5d.1.B | 20.75 | hetero-oligomer | HHblits | X-ray | 4.80Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 3a5d.2.C | 20.75 | hetero-oligomer | HHblits | X-ray | 4.80Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 1fq0.1.A | 17.86 | homo-trimer | HHblits | X-ray | 2.10Å | 0.27 | 0.21 | KDPG ALDOLASE |
| 1eua.1.C | 17.86 | homo-trimer | HHblits | X-ray | 1.95Å | 0.27 | 0.21 | KDPG ALDOLASE |
| 3qvj.1.A | 20.00 | homo-hexamer | HHblits | X-ray | 2.10Å | 0.28 | 0.21 | Putative hydantoin racemase |
| 3nd9.1.A | 21.15 | monomer | HHblits | X-ray | 3.10Å | 0.32 | 0.20 | V-type ATP synthase alpha chain |
| 4mwa.1.A | 14.81 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.1.B | 14.81 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.1.C | 14.81 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.1.D | 14.81 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.2.B | 14.81 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 4mwa.2.D | 14.81 | homo-tetramer | HHblits | X-ray | 1.85Å | 0.29 | 0.20 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 2f6u.1.A | 20.75 | homo-dimer | HHblits | X-ray | 1.55Å | 0.30 | 0.20 | (S)-3-O-Geranylgeranylglyceryl Phosphate Synthase |
| 2f6x.1.A | 20.75 | homo-dimer | HHblits | X-ray | 2.00Å | 0.30 | 0.20 | (S)-3-O-Geranylgeranylglyceryl Phosphate Synthase |
| 1yht.1.A | 15.09 | monomer | HHblits | X-ray | 2.00Å | 0.29 | 0.20 | DspB |
| 3bol.1.A | 21.15 | homo-dimer | HHblits | X-ray | 1.85Å | 0.30 | 0.20 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 1q7m.2.A | 21.15 | monomer | HHblits | X-ray | 2.10Å | 0.30 | 0.20 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 1q7q.1.A | 21.15 | monomer | HHblits | X-ray | 3.10Å | 0.30 | 0.20 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 1q8a.2.A | 21.15 | monomer | HHblits | X-ray | 1.70Å | 0.30 | 0.20 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 4uhc.1.A | 17.65 | monomer | HHblits | X-ray | 1.03Å | 0.32 | 0.19 | ESTERASE |
| 4uhd.1.A | 17.65 | monomer | HHblits | X-ray | 1.07Å | 0.32 | 0.19 | ESTERASE |
| 1fq0.1.A | 17.31 | homo-trimer | HHblits | X-ray | 2.10Å | 0.29 | 0.20 | KDPG ALDOLASE |
| 1eua.1.C | 17.31 | homo-trimer | HHblits | X-ray | 1.95Å | 0.29 | 0.20 | KDPG ALDOLASE |
| 4fq5.1.A | 13.21 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.20 | Maleate cis-trans isomerase |
| 4fq5.1.B | 13.21 | homo-tetramer | HHblits | X-ray | 2.10Å | 0.27 | 0.20 | Maleate cis-trans isomerase |
| 4fq7.1.A | 13.46 | homo-tetramer | HHblits | X-ray | 3.00Å | 0.28 | 0.20 | Maleate cis-trans isomerase |
| 4mg4.1.A | 23.53 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.19 | Phosphonomutase |
| 4mg4.2.B | 23.53 | homo-dimer | HHblits | X-ray | 1.70Å | 0.29 | 0.19 | Phosphonomutase |
| 1xuu.1.A | 22.92 | homo-dimer | HHblits | X-ray | 1.90Å | 0.33 | 0.18 | polysialic acid capsule biosynthesis protein SiaC |
| 4ih4.1.A | 16.00 | monomer | HHblits | X-ray | 3.50Å | 0.29 | 0.19 | AT3g03990/T11I18_10 |
| 5hzg.2.A | 16.00 | hetero-oligomer | HHblits | X-ray | 3.30Å | 0.29 | 0.19 | Strigolactone esterase D14 |
| 4dnh.1.A | 12.00 | monomer | HHblits | X-ray | 2.50Å | 0.29 | 0.19 | Uncharacterized protein |
| 2wqp.1.A | 23.40 | homo-dimer | HHblits | X-ray | 1.75Å | 0.33 | 0.18 | POLYSIALIC ACID CAPSULE BIOSYNTHESIS PROTEIN SIAC |
| 1wbh.1.A | 20.00 | homo-trimer | HHblits | X-ray | 1.55Å | 0.28 | 0.19 | KHG/KDPG ALDOLASE |
| 1fwr.1.A | 20.00 | homo-trimer | HHblits | X-ray | 2.70Å | 0.28 | 0.19 | KDPG ALDOLASE |
| 5y2x.1.A | 11.76 | monomer | HHblits | X-ray | 2.02Å | 0.26 | 0.19 | Haloalkane dehalogenase |
| 5y2y.2.A | 11.76 | monomer | HHblits | X-ray | 2.27Å | 0.26 | 0.19 | Haloalkane dehalogenase |
| 1fui.1.A | 11.76 | homo-trimer | HHblits | X-ray | 2.50Å | 0.25 | 0.19 | L-FUCOSE ISOMERASE |
| 4c22.1.A | 8.00 | homo-hexamer | HHblits | X-ray | 2.70Å | 0.25 | 0.19 | L-FUCOSE ISOMERASE |
| 4c20.1.A | 6.00 | monomer | HHblits | X-ray | 2.41Å | 0.25 | 0.19 | L-FUCOSE ISOMERASE |
| 4c21.1.A | 6.00 | homo-hexamer | HHblits | X-ray | 2.55Å | 0.25 | 0.19 | L-FUCOSE ISOMERASE |
| 2ip4.1.A | 16.67 | monomer | HHblits | X-ray | 2.80Å | 0.28 | 0.18 | Phosphoribosylamine--glycine ligase |
| 2ip4.2.A | 16.67 | monomer | HHblits | X-ray | 2.80Å | 0.28 | 0.18 | Phosphoribosylamine--glycine ligase |
| 1rdu.1.A | 17.02 | monomer | HHblits | NMR | NA | 0.29 | 0.18 | conserved hypothetical protein |
| 3a9t.1.A | 12.24 | homo-hexamer | HHblits | X-ray | 2.61Å | 0.25 | 0.18 | D-arabinose isomerase |
| 4ix1.1.A | 8.16 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.18 | hypothetical protein |
| 4ix1.1.B | 8.16 | homo-dimer | HHblits | X-ray | 2.80Å | 0.25 | 0.18 | hypothetical protein |
| 1ulz.1.A | 19.15 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.18 | pyruvate carboxylase n-terminal domain |
| 5vev.1.A | 8.33 | monomer | HHblits | X-ray | 1.90Å | 0.25 | 0.18 | Phosphoribosylamine--glycine ligase |
| 4h4x.1.A | 30.95 | homo-dimer | HHblits | X-ray | 1.50Å | 0.34 | 0.16 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4fdc.1.A | 29.27 | monomer | HHblits | X-ray | 2.40Å | 0.35 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 5fs9.1.A | 29.27 | monomer | HHblits | X-ray | 1.75Å | 0.35 | 0.15 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs6.1.A | 29.27 | monomer | HHblits | X-ray | 1.90Å | 0.35 | 0.15 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fmh.1.A | 29.27 | monomer | HHblits | X-ray | 1.80Å | 0.35 | 0.15 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs8.1.A | 29.27 | monomer | HHblits | X-ray | 1.40Å | 0.35 | 0.15 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 4qwv.1.A | 9.09 | monomer | HHblits | X-ray | 2.45Å | 0.30 | 0.17 | High-affinity leucine-specific transport system periplasmic binding protein, Chemotaxis protein CheY |
| 4h4v.1.A | 31.71 | homo-dimer | HHblits | X-ray | 1.40Å | 0.35 | 0.15 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 4h4t.1.A | 31.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.35 | 0.15 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 2yvg.1.A | 31.71 | homo-dimer | HHblits | X-ray | 1.60Å | 0.35 | 0.15 | Ferredoxin reductase |
| 2yvj.1.A | 31.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.35 | 0.15 | Ferredoxin reductase |
| 2yvj.1.C | 31.71 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.35 | 0.15 | Ferredoxin reductase |
| 1d7y.1.A | 31.71 | homo-dimer | HHblits | X-ray | 2.10Å | 0.35 | 0.15 | FERREDOXIN REDUCTASE |
| 4h4z.1.A | 31.71 | homo-dimer | HHblits | X-ray | 1.95Å | 0.35 | 0.15 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 2gr3.1.A | 31.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.35 | 0.15 | ferredoxin reductase |
| 4h4p.1.A | 31.71 | homo-dimer | HHblits | X-ray | 1.50Å | 0.35 | 0.15 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 2yya.1.A | 10.87 | monomer | HHblits | X-ray | 2.40Å | 0.25 | 0.17 | Phosphoribosylamine--glycine ligase |
| 3ef6.1.A | 27.50 | monomer | HHblits | X-ray | 1.80Å | 0.36 | 0.15 | Toluene 1,2-dioxygenase system ferredoxin--NAD(+) reductase |
| 4emi.1.A | 27.50 | monomer | HHblits | X-ray | 1.81Å | 0.36 | 0.15 | TodA |
| 4emj.1.A | 27.50 | hetero-oligomer | HHblits | X-ray | 2.40Å | 0.36 | 0.15 | TodA |
| 4bur.1.A | 30.00 | monomer | HHblits | X-ray | 2.88Å | 0.36 | 0.15 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 4bur.3.A | 30.00 | monomer | HHblits | X-ray | 2.88Å | 0.36 | 0.15 | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL |
| 5fs7.1.A | 30.00 | monomer | HHblits | X-ray | 1.85Å | 0.36 | 0.15 | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL |
| 1m6i.1.A | 30.00 | monomer | HHblits | X-ray | 1.80Å | 0.36 | 0.15 | Programmed cell death protein 8 |
| 4lii.1.A | 30.00 | monomer | HHblits | X-ray | 1.88Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 1gv4.1.A | 30.00 | monomer | HHblits | X-ray | 2.00Å | 0.36 | 0.15 | PROGRAMED CELL DEATH PROTEIN 8 |
| 5kvi.1.A | 30.00 | monomer | HHblits | X-ray | 2.00Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd4.1.A | 30.00 | homo-dimer | HHblits | X-ray | 2.24Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd4.1.B | 30.00 | homo-dimer | HHblits | X-ray | 2.24Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 5kvh.1.A | 30.00 | homo-dimer | HHblits | X-ray | 2.27Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.1.A | 30.00 | monomer | HHblits | X-ray | 2.95Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.2.A | 30.00 | monomer | HHblits | X-ray | 2.95Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 3gd3.4.A | 30.00 | monomer | HHblits | X-ray | 2.95Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 5miu.1.A | 30.00 | monomer | HHblits | X-ray | 3.50Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.A | 30.00 | homo-dimer | HHblits | X-ray | 3.10Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 5miv.1.B | 30.00 | homo-dimer | HHblits | X-ray | 3.10Å | 0.36 | 0.15 | Apoptosis-inducing factor 1, mitochondrial |
| 5jcm.1.A | 18.60 | monomer | HHblits | X-ray | 1.90Å | 0.30 | 0.16 | Os09g0567300 protein |
| 4naf.1.A | 23.81 | hetero-oligomer | HHblits | X-ray | 1.90Å | 0.31 | 0.16 | Heptaprenylglyceryl phosphate synthase |
| 4h4r.1.A | 32.50 | homo-dimer | HHblits | X-ray | 1.40Å | 0.35 | 0.15 | Biphenyl dioxygenase ferredoxin reductase subunit |
| 5jcl.1.A | 18.60 | monomer | HHblits | X-ray | 1.80Å | 0.29 | 0.16 | Os09g0567300 protein |
| 3lxd.1.A | 21.43 | monomer | HHblits | X-ray | 2.50Å | 0.31 | 0.16 | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| 3fg2.1.A | 21.95 | monomer | HHblits | X-ray | 2.20Å | 0.32 | 0.15 | Putative rubredoxin reductase |
| 2v3a.1.A | 24.39 | monomer | HHblits | X-ray | 2.40Å | 0.32 | 0.15 | RUBREDOXIN REDUCTASE |
| 2v3b.1.A | 24.39 | hetero-oligomer | HHblits | X-ray | 2.45Å | 0.32 | 0.15 | RUBREDOXIN REDUCTASE |
| 4fx9.1.A | 14.29 | homo-dimer | HHblits | X-ray | 2.70Å | 0.29 | 0.16 | Coenzyme A disulfide reductase |
| 3ics.1.A | 17.07 | homo-dimer | HHblits | X-ray | 1.94Å | 0.31 | 0.15 | Coenzyme A-Disulfide Reductase |
| 3lb8.1.A | 21.95 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.31 | 0.15 | Putidaredoxin reductase |
| 3lb8.2.A | 21.95 | hetero-oligomer | HHblits | X-ray | 2.60Å | 0.31 | 0.15 | Putidaredoxin reductase |
| 2y85.1.A | 19.05 | monomer | HHblits | X-ray | 2.40Å | 0.29 | 0.16 | PHOSPHORIBOSYL ISOMERASE A |
| 3zs4.1.A | 19.05 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.16 | PHOSPHORIBOSYL ISOMERASE A |
| 3ga5.1.A | 9.30 | monomer | HHblits | X-ray | 1.87Å | 0.27 | 0.16 | D-galactose-binding periplasmic protein |
| 1q1r.1.A | 21.95 | monomer | HHblits | X-ray | 1.91Å | 0.30 | 0.15 | Putidaredoxin reductase |
| 1q1w.2.A | 21.95 | monomer | HHblits | X-ray | 2.60Å | 0.30 | 0.15 | Putidaredoxin reductase |
| 3hyv.1.A | 23.08 | homo-trimer | HHblits | X-ray | 2.30Å | 0.34 | 0.15 | Sulfide-quinone reductase |
| 2gps.1.A | 11.36 | monomer | HHblits | X-ray | 2.80Å | 0.24 | 0.17 | Biotin carboxylase |
| 2o14.1.A | 17.07 | monomer | HHblits | X-ray | 2.10Å | 0.28 | 0.15 | Hypothetical protein yxiM |
| 5ere.1.A | 19.51 | monomer | HHblits | X-ray | 2.00Å | 0.28 | 0.15 | Extracellular ligand-binding receptor |
| 3rv3.1.A | 11.63 | homo-dimer | HHblits | X-ray | 1.91Å | 0.24 | 0.16 | Biotin carboxylase |
| 4fx7.1.A | 17.07 | monomer | HHblits | X-ray | 2.08Å | 0.28 | 0.15 | Imidazole glycerol phosphate synthase subunit HisF |
| 3kd9.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.75Å | 0.30 | 0.15 | Coenzyme A disulfide reductase |
| 5l1n.1.A | 15.00 | homo-dimer | HHblits | X-ray | 3.60Å | 0.30 | 0.15 | Coenzyme A disulfide reductase |
| 3ipc.1.A | 15.00 | monomer | HHblits | X-ray | 1.30Å | 0.29 | 0.15 | ABC transporter, substrate binding protein (Amino acid) |
| 3cge.1.A | 9.76 | homo-dimer | HHblits | X-ray | 2.26Å | 0.27 | 0.15 | Pyridine nucleotide-disulfide oxidoreductase, class I |
| 4em3.1.A | 12.50 | homo-dimer | HHblits | X-ray | 1.98Å | 0.29 | 0.15 | Coenzyme A disulfide reductase |
| 2c7g.1.A | 9.76 | homo-dimer | HHblits | X-ray | 1.80Å | 0.27 | 0.15 | NADPH-FERREDOXIN REDUCTASE FPRA |
| 3zfh.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.28 | 0.15 | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE |
| 1usi.1.A | 10.26 | monomer | HHblits | X-ray | 1.80Å | 0.30 | 0.15 | LEUCINE-SPECIFIC BINDING PROTEIN |
| 1usg.1.A | 10.26 | homo-dimer | HHblits | X-ray | 1.53Å | 0.30 | 0.15 | LEUCINE-SPECIFIC BINDING PROTEIN |
| 4dqw.1.A | 12.50 | homo-octamer | HHblits | X-ray | 2.51Å | 0.28 | 0.15 | Inosine-5'-monophosphate dehydrogenase |
| 4dqw.1.B | 12.50 | homo-octamer | HHblits | X-ray | 2.51Å | 0.28 | 0.15 | Inosine-5'-monophosphate dehydrogenase |
| 2liv.1.A | 10.26 | monomer | HHblits | X-ray | 2.40Å | 0.30 | 0.15 | LEUCINE |
| 3noy.1.A | 20.51 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.15 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 3noy.2.A | 20.51 | homo-dimer | HHblits | X-ray | 2.70Å | 0.30 | 0.15 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| 3cgb.1.A | 10.00 | homo-dimer | HHblits | X-ray | 1.90Å | 0.27 | 0.15 | Pyridine nucleotide-disulfide oxidoreductase, class I |
| 3ipa.1.A | 15.38 | monomer | HHblits | X-ray | 1.55Å | 0.29 | 0.15 | ABC transporter, substrate binding protein (Amino acid) |
| 1lqu.1.A | 10.00 | monomer | HHblits | X-ray | 1.25Å | 0.27 | 0.15 | FprA |
| 4eqr.1.A | 12.82 | homo-dimer | HHblits | X-ray | 1.80Å | 0.29 | 0.15 | Coenzyme A disulfide reductase |
| 4nas.1.A | 29.73 | homo-dimer | HHblits | X-ray | 1.92Å | 0.33 | 0.14 | Ribulose-bisphosphate carboxylase |
| 1x7i.1.A | 18.42 | homo-dimer | HHblits | X-ray | 1.70Å | 0.31 | 0.14 | Copper homeostasis protein cutC |
| 1x7i.1.B | 18.42 | homo-dimer | HHblits | X-ray | 1.70Å | 0.31 | 0.14 | Copper homeostasis protein cutC |
| 1twd.1.A | 18.42 | homo-dimer | HHblits | X-ray | 1.70Å | 0.31 | 0.14 | Copper homeostasis protein cutC |
| 1twd.1.B | 18.42 | homo-dimer | HHblits | X-ray | 1.70Å | 0.31 | 0.14 | Copper homeostasis protein cutC |
| 4kv7.1.A | 23.08 | monomer | HHblits | X-ray | 1.20Å | 0.28 | 0.15 | Probable leucine/isoleucine/valine-binding protein |
| 1e1m.1.A | 15.38 | monomer | HHblits | X-ray | 1.85Å | 0.28 | 0.15 | ADRENODOXIN REDUCTASE |
| 1e1l.1.A | 15.38 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | ADRENODOXIN REDUCTASE |
| 1e6e.2.A | 15.38 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | NADPH\:ADRENODOXIN OXIDOREDUCTASE |
| 1e6e.1.A | 15.38 | hetero-oligomer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | NADPH\:ADRENODOXIN OXIDOREDUCTASE |
| 4n0q.1.A | 17.95 | monomer | HHblits | X-ray | 2.30Å | 0.28 | 0.15 | Leu/Ile/Val-binding protein homolog 3 |
| 3a5d.1.A | 18.42 | hetero-oligomer | HHblits | X-ray | 4.80Å | 0.30 | 0.14 | V-type ATP synthase alpha chain |
| 3a5d.1.B | 18.42 | hetero-oligomer | HHblits | X-ray | 4.80Å | 0.30 | 0.14 | V-type ATP synthase alpha chain |
| 3a5d.2.C | 18.42 | hetero-oligomer | HHblits | X-ray | 4.80Å | 0.30 | 0.14 | V-type ATP synthase alpha chain |
| 2czw.1.A | 15.79 | monomer | HHblits | X-ray | 1.90Å | 0.29 | 0.14 | 50S ribosomal protein L7Ae |
| 2oek.1.A | 27.03 | homo-dimer | HHblits | X-ray | 1.80Å | 0.31 | 0.14 | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase |
| 4lck.1.A | 12.82 | monomer | HHblits | X-ray | 3.20Å | 0.27 | 0.15 | Ribosomal protein YbxF |
| 4lck.2.A | 12.82 | monomer | HHblits | X-ray | 3.20Å | 0.27 | 0.15 | Ribosomal protein YbxF |
| 2zvi.1.A | 27.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.14 | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase |
| 2zvi.1.B | 27.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.14 | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase |
| 2zvi.2.A | 27.03 | homo-dimer | HHblits | X-ray | 2.30Å | 0.30 | 0.14 | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase |
| 4n0q.1.A | 15.79 | monomer | HHblits | X-ray | 2.30Å | 0.27 | 0.14 | Leu/Ile/Val-binding protein homolog 3 |
| 4nut.1.A | 18.42 | hetero-oligomer | HHblits | X-ray | 1.55Å | 0.27 | 0.14 | 13 kDa ribonucleoprotein-associated protein |
| 3jcm.1.G | 18.42 | hetero-oligomer | HHblits | EM | NA | 0.27 | 0.14 | 13 kDa ribonucleoprotein-associated protein |
| 1zwz.1.A | 18.42 | monomer | HHblits | X-ray | 1.90Å | 0.27 | 0.14 | Snu13p |
| 5wyk.1.G | 18.42 | hetero-oligomer | HHblits | EM | NA | 0.27 | 0.14 | 13 kDa ribonucleoprotein-associated protein |
| 5wyj.7.A | 18.42 | monomer | HHblits | EM | NA | 0.27 | 0.14 | 13 kDa ribonucleoprotein-associated protein |
| 5nrl.1.R | 18.42 | hetero-oligomer | HHblits | EM | NA | 0.27 | 0.14 | 13 kDa ribonucleoprotein-associated protein |
| 5wlc.48.A | 18.42 | monomer | HHblits | EM | NA | 0.27 | 0.14 | Snu13 |
| 5wlc.47.A | 18.42 | monomer | HHblits | EM | NA | 0.27 | 0.14 | Snu13 |
| 4kv7.1.A | 15.38 | monomer | HHblits | X-ray | 1.20Å | 0.25 | 0.15 | Probable leucine/isoleucine/valine-binding protein |
| 3vrd.1.B | 22.22 | hetero-oligomer | HHblits | X-ray | 1.50Å | 0.31 | 0.14 | Flavocytochrome c flavin subunit |
| 3ipa.1.A | 13.16 | monomer | HHblits | X-ray | 1.55Å | 0.27 | 0.14 | ABC transporter, substrate binding protein (Amino acid) |
| 2hqb.1.A | 5.13 | monomer | HHblits | X-ray | 2.70Å | 0.24 | 0.15 | Transcriptional activator of comK gene |
| 4f06.1.A | 10.53 | monomer | HHblits | X-ray | 1.30Å | 0.26 | 0.14 | Extracellular ligand-binding receptor |
| 3lkv.1.A | 16.22 | homo-dimer | HHblits | X-ray | 2.20Å | 0.28 | 0.14 | uncharacterized CONSERVED DOMAIN PROTEIN |
| 3ipc.1.A | 13.89 | monomer | HHblits | X-ray | 1.30Å | 0.27 | 0.14 | ABC transporter, substrate binding protein (Amino acid) |
| 2lbp.1.A | 5.26 | monomer | HHblits | X-ray | 2.40Å | 0.22 | 0.14 | LEUCINE-BINDING PROTEIN |
| 3hly.1.A | 20.00 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.13 | Flavodoxin-like domain |
| 3hly.2.A | 20.00 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.13 | Flavodoxin-like domain |
| 3hly.3.A | 20.00 | monomer | HHblits | X-ray | 2.40Å | 0.28 | 0.13 | Flavodoxin-like domain |
| 4evs.1.A | 11.43 | monomer | HHblits | X-ray | 1.45Å | 0.27 | 0.13 | PUTATIVE ABC TRANSPORTER SUBUNIT, SUBSTRATE-BINDING COMPONENT |
| 3ijd.1.A | 17.65 | homo-dimer | HHblits | X-ray | 2.00Å | 0.28 | 0.13 | uncharacterized protein |
| 1usi.1.A | 5.56 | monomer | HHblits | X-ray | 1.80Å | 0.23 | 0.14 | LEUCINE-SPECIFIC BINDING PROTEIN |
| 1usg.1.A | 5.56 | homo-dimer | HHblits | X-ray | 1.53Å | 0.23 | 0.14 | LEUCINE-SPECIFIC BINDING PROTEIN |
| 4mld.1.A | 15.15 | homo-dimer | HHblits | X-ray | 2.88Å | 0.30 | 0.12 | Response regulator |
| 4mld.1.B | 15.15 | homo-dimer | HHblits | X-ray | 2.88Å | 0.30 | 0.12 | Response regulator |
| 4mld.2.A | 15.15 | homo-dimer | HHblits | X-ray | 2.88Å | 0.30 | 0.12 | Response regulator |
| 4mld.2.B | 15.15 | homo-dimer | HHblits | X-ray | 2.88Å | 0.30 | 0.12 | Response regulator |
| 1z16.1.A | 5.56 | monomer | HHblits | X-ray | 1.72Å | 0.23 | 0.14 | Leu/Ile/Val-binding protein |
| 1z15.1.A | 5.56 | monomer | HHblits | X-ray | 1.70Å | 0.23 | 0.14 | Leu/Ile/Val-binding protein |
| 3e61.1.A | 12.50 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.29 | 0.12 | Putative transcriptional repressor of ribose operon |
| 3qg1.1.A | 26.67 | monomer | HHblits | X-ray | 2.95Å | 0.34 | 0.11 | V-type ATP synthase alpha chain |
| 5dmm.1.A | 12.50 | monomer | HHblits | X-ray | 1.78Å | 0.25 | 0.12 | Homocysteine S-methyltransferase |
| 5dmn.1.A | 12.50 | homo-dimer | HHblits | X-ray | 2.89Å | 0.25 | 0.12 | Homocysteine S-methyltransferase |
| 3dfu.1.A | 25.93 | homo-dimer | HHblits | X-ray | 2.07Å | 0.34 | 0.10 | Uncharacterized Protein from 6-Phosphogluconate Dehydrogenase-Like Family |
| 3dfu.1.B | 25.93 | homo-dimer | HHblits | X-ray | 2.07Å | 0.34 | 0.10 | Uncharacterized Protein from 6-Phosphogluconate Dehydrogenase-Like Family |
| 5a9r.1.A | 32.00 | homo-dimer | HHblits | X-ray | 1.55Å | 0.36 | 0.09 | IMINE REDUCTASE |
| 5a9s.1.A | 32.00 | hetero-oligomer | HHblits | X-ray | 2.06Å | 0.36 | 0.09 | IMINE REDUCTASE |
| 5fwn.1.B | 32.00 | homo-dimer | HHblits | X-ray | 2.14Å | 0.36 | 0.09 | IMINE REDUCTASE |
| 5a9s.1.B | 32.00 | hetero-oligomer | HHblits | X-ray | 2.06Å | 0.36 | 0.09 | IMINE REDUCTASE |
| 3bol.1.A | 32.00 | homo-dimer | HHblits | X-ray | 1.85Å | 0.35 | 0.09 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 1q7m.2.A | 32.00 | monomer | HHblits | X-ray | 2.10Å | 0.35 | 0.09 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 1q7q.1.A | 32.00 | monomer | HHblits | X-ray | 3.10Å | 0.35 | 0.09 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 1q8a.2.A | 32.00 | monomer | HHblits | X-ray | 1.70Å | 0.35 | 0.09 | 5-methyltetrahydrofolate S-homocysteine methyltransferase |
| 3lkv.1.A | 18.52 | homo-dimer | HHblits | X-ray | 2.20Å | 0.27 | 0.10 | uncharacterized CONSERVED DOMAIN PROTEIN |
| 3pn9.1.A | 11.54 | homo-tetramer | HHblits | X-ray | 2.00Å | 0.27 | 0.10 | Proline dipeptidase |
| 4rs3.1.A | 16.67 | monomer | HHblits | X-ray | 1.40Å | 0.31 | 0.09 | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein |
| 3p94.1.A | 8.00 | monomer | HHblits | X-ray | 1.93Å | 0.26 | 0.09 | GDSL-like Lipase |
| 3iwp.1.B | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.1.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.2.B | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.2.C | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.2.D | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.3.A | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.3.B | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.3.C | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 3iwp.3.D | 16.67 | homo-tetramer | HHblits | X-ray | 2.50Å | 0.29 | 0.09 | Copper homeostasis protein cutC homolog |
| 5kp9.1.A | 17.39 | homo-60-mer | HHblits | EM | NA | 0.31 | 0.09 | EPN-01* |
| 1drk.1.A | 12.50 | monomer | HHblits | X-ray | 2.00Å | 0.26 | 0.09 | D-RIBOSE-BINDING PROTEIN |
| 2iz1.1.B | 28.57 | homo-dimer | HHblits | X-ray | 2.30Å | 0.36 | 0.08 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 2iz1.1.A | 28.57 | homo-dimer | HHblits | X-ray | 2.30Å | 0.36 | 0.08 | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING |
| 1glg.1.A | 13.04 | monomer | HHblits | X-ray | 2.00Å | 0.27 | 0.09 | GALACTOSE/GLUCOSE-BINDING PROTEIN |
| 2qw1.1.A | 13.04 | monomer | HHblits | X-ray | 1.70Å | 0.27 | 0.09 | D-galactose-binding periplasmic protein |
| 2fvy.1.A | 13.04 | monomer | HHblits | X-ray | 0.92Å | 0.27 | 0.09 | D-galactose-binding periplasmic protein |
| 2ipm.1.A | 13.04 | monomer | HHblits | X-ray | 1.12Å | 0.27 | 0.09 | D-galactose-binding periplasmic protein |
| 2ipn.1.A | 13.04 | monomer | HHblits | X-ray | 1.15Å | 0.27 | 0.09 | D-galactose-binding periplasmic protein |
| 2ipl.1.A | 13.64 | monomer | HHblits | X-ray | 1.20Å | 0.28 | 0.08 | D-galactose-binding periplasmic protein |
| 5kws.1.A | 13.64 | monomer | HHblits | X-ray | 1.32Å | 0.28 | 0.08 | Galactose-binding protein |
| 2hph.1.A | 13.64 | monomer | HHblits | X-ray | 1.33Å | 0.28 | 0.08 | D-galactose-binding periplasmic protein |
| 3tq0.1.A | 14.29 | homo-dimer | HHblits | X-ray | 1.90Å | 0.28 | 0.08 | Dihydroorotate dehydrogenase |
| 3pid.1.A | 30.00 | homo-dimer | HHblits | X-ray | 1.40Å | 0.31 | 0.08 | UDP-glucose 6-dehydrogenase |
| 2liv.1.A | 15.00 | monomer | HHblits | X-ray | 2.40Å | 0.30 | 0.08 | LEUCINE |
| 3bbl.1.A | 15.00 | homo-dimer | HHblits | X-ray | 2.35Å | 0.26 | 0.08 | Regulatory protein of LacI family |
| 3k9c.1.A | 16.67 | homo-dimer | HHblits | X-ray | 2.14Å | 0.30 | 0.07 | Transcriptional regulator, LacI family protein |
| 4dqw.1.A | 31.25 | homo-octamer | HHblits | X-ray | 2.51Å | 0.33 | 0.06 | Inosine-5'-monophosphate dehydrogenase |
| 4dqw.1.B | 31.25 | homo-octamer | HHblits | X-ray | 2.51Å | 0.33 | 0.06 | Inosine-5'-monophosphate dehydrogenase |
| 3zfh.1.A | 31.25 | homo-tetramer | HHblits | X-ray | 2.25Å | 0.33 | 0.06 | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE |